Abstract
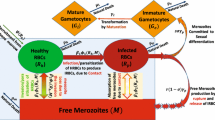
Mathematical modeling provides a rigorous way to quantify immunological processes and discriminate between alternative mechanisms driving specific biological phenomena. It is typical that mathematical models of immunological phenomena are developed by modelers to explain specific sets of experimental data after the data have been collected by experimental collaborators. Whether the available data are sufficient to accurately estimate model parameters or to discriminate between alternative models is not typically investigated. While previously collected data may be sufficient to guide development of alternative models and help estimating model parameters, such data often do not allow to discriminate between alternative models. As a case study, we develop a series of power analyses to determine optimal sample sizes that allow for accurate estimation of model parameters and for discrimination between alternative models describing clustering of CD8 T cells around Plasmodium liver stages. In our typical experiments, mice are infected intravenously with Plasmodium sporozoites that invade hepatocytes (liver cells), and then activated CD8 T cells are transferred into the infected mice. The number of T cells found in the vicinity of individual infected hepatocytes at different times after T cell transfer is counted using intravital microscopy. We previously developed a series of mathematical models aimed to explain highly variable number of T cells per parasite; one of such models, the density-dependent recruitment (DDR) model, fitted the data from preliminary experiments better than the alternative models, such as the density-independent exit (DIE) model. Here, we show that the ability to discriminate between these alternative models depends on the number of parasites imaged in the analysis; analysis of about \(n=50\) parasites at 2, 4, and 8 h after T cell transfer will allow for over 95% probability to select the correct model. The type of data collected also has an impact; following T cell clustering around individual parasites over time (called as longitudinal (LT) data) allows for a more precise and less biased estimates of the parameters of the DDR model than that generated from a more traditional way of imaging individual parasites in different liver areas/mice (cross-sectional (CS) data). However, LT imaging comes at a cost of a need to keep the mice alive under the microscope for hours which may be ethically unacceptable. We finally show that the number of time points at which the measurements are taken also impacts the precision of estimation of DDR model parameters; in particular, measuring T cell clustering at one time point does not allow accurately estimating all parameters of the DDR model. Using our case study, we propose a general framework on how mathematical modeling can be used to guide experimental designs and power analyses of complex biological processes.





Similar content being viewed by others
Code Availability
Simulations have been performed either in MATLAB (version 2018b) or in Mathematica (version 11.2 or 12.3). Simulation codes will be made available upon request.
Abbreviations
- DDR:
-
Density-dependent recruitment
- DIE:
-
Density-independent exit
- LT:
-
Longitudinal
- CS:
-
Cross-sectional
- LTL:
-
Longitudinal likelihood
- CSL:
-
Cross-sectional likelihood
- NBD:
-
Negative binomial distribution
References
Akbari M, Kimura K, Bayarsaikhan G, Kimura D, Miyakoda M, Juriasingani S, Yuda M, Amino R, Yui K (2018) Nonspecific CD8+ T cells and dendritic cells/macrophages participate in formation of CD8+ T cell-mediated clusters against malaria liver-stage infection. Infect Immun 86(4):e00-717–17
Bailey NTJ (1964) The elements of stochastic processes with applications to the natural sciences. Wiley, New York
Bera S, Amino R, Cockburn IA, Ganusov VV (2022) Mathematical modeling suggests that a single Plasmodium-specific CD8 T cell can eliminate a malaria liver stage in mice. BioRxiv 1:1–44. https://doi.org/10.1101/2022.05.18.492520
Braeckel-Budimir NV, Harty JT (2014) CD8 T-cell-mediated protection against liver-stage malaria: lessons from a mouse model. Front Microbiol 5:272. https://doi.org/10.3389/fmicb.2014.00272
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach. Springer, New York
Cobelli C, DiStefano JJ (1980) Parameter and structural identifiability concepts and ambiguities: a critical review and analysis. Am J Physiol 239:R7-24. https://doi.org/10.1152/ajpregu.1980.239.1.R7
Cockburn IA, Amino R, Kelemen RK, Kuo SC, Tse S-W, Radtke A, Mac-Daniel L, Ganusov VV, Zavala F et al (2013) In vivo imaging of CD8+ T cell-mediated elimination of malaria liver stages. Proc Natl Acad Sci 110(22):9090–9095
Ganusov VV (2016) Strong inference in mathematical modeling: a method for robust science in the twenty-first century. Front Microbiol 7:1131. https://doi.org/10.3389/fmicb.2016.01131
Genser B, Cooper PJ, Yazdanbakhsh M, Barreto ML, Rodrigues LC (2007) A guide to modern statistical analysis of immunological data. BMC Immunol 8:27. https://doi.org/10.1186/1471-2172-8-27
Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81(25):2340–2361
Goel NS, Richter-Dyn N (2016) Stochastic models in biology. Elsevier
Hoft SG, Sallin MA, Kauffman KD, Sakai S, Ganusov VV, Barber DL (2019) The rate of CD4 T cell entry into the lungs during Mycobacterium tuberculosis infection is determined by partial and opposing effects of multiple chemokine receptors. Infect Immun 87(e00):841–918. https://doi.org/10.1128/IAI.00841-18
Kelemen RK, Rajakaruna H, Cockburn IA, Ganu sov VV (2019) Clustering of activated CD8 T cells around malaria-infected hepat ocytes is rapid and is driven by antigen-specific cells. Front Immunol 10:2153. https://doi.org/10.3389/fimmu.2019.02153
Kimura K, Kimura D, Matsushima Y, Miyakoda M, Honma K, Yuda M, Yui K (2013) CD8+ T cells specific for a malaria cytoplasmic antigen form clusters around infected hepatocytes and are protective at the liver stage of infection. Infect Immun 81:3825–3834. https://doi.org/10.1128/IAI.00570-13
Lefebvre MN, Surette FA, Anthony SM, Vijay R, Jensen IJ, Pewe LL, Hancox LS, Van Braeckel-Budimir N, van de Wall S et al (2021) Expeditious recruitment of circulating memory CD8 T cells to the liver facilitates control of malaria. Cell reports 37(109):956. https://doi.org/10.1016/j.celrep.2021.109956
McDaniel MM, Krishna N, Handagama WG, Eda S, Ganusov VV (2016) Quantifying limits on replication, death, and quiescence of Mycobacterium tuberculosis in mice. Front Microbiol 7:862. https://doi.org/10.3389/fmicb.2016.00862
Miller J, Burch-Smith TM, Ganusov VV (2022) Mathematical modeling suggests cooperation of plant-infecting viruses. Viruses 14(4):1. https://doi.org/10.3390/v14040741
Parthasarathy P (1987) A transient solution to an M/M/1 queue: a simple approach. Adv Appl Probab 19(4):997–998
Renshaw E (2011) Stochastic population processes: analysis, approximations, simulations. Oxford University Press. https://doi.org/10.1093/acprof:oso/9780199575312.001.0001.
Schmidt NW, Podyminogin RL, Butler NS, Badovinac VP, Tucker BJ, Bahjat KS, Lauer P, Reyes-Sandoval A, Hutchings CL et al (2008) Memory CD8 T cell responses exceeding a large but definable threshold provide long-term immunity to malaria. Proc Natl Acad Sci USA 105(37):14-017–22
Schmidt NW, Butler NS, Harty JT (2011) Plasmodium-host interactions directly influence the threshold of memory CD8 T cells required for protective immunity. J Immunol 186(10):5873–5884. https://doi.org/10.4049/jimmunol.1100194
Shin SC, Vanderberg JP, Terzakis JA (1982) Direct infection of hepatocytes by sporozoites of Plasmodium berghei. J Protozool 29:448–454. https://doi.org/10.1111/j.1550-7408.1982.tb05431.x
Sinnis P, Willnow TE, Briones MR, Herz J, Nussenzweig V (1996) Remnant lipoproteins inhibit malaria sporozoite invasion of hepatocytes. J Experim Med 184:945–954. https://doi.org/10.1084/jem.184.3.945
Sokal RR, Rohlf FJ (1987) Biostatistics. Francise & Co, New York
Song H, Giorgi EE, Ganusov VV, Cai FN, Athreya G, Yoon H, Carja O, Hora B, Hraber P et al (2018) Tracking HIV-1 recombination to resolve its contribution to HIV-1 evolution in natural infection. Nat Commun 9:1928. https://doi.org/10.1038/s41467-018-04217-5
Zenkov VS, O’Connor J, Cockburn IA, Ganusov VV (2022) A new method based on the von Mises–Fisher distribution shows that a minority of liver-localized CD8 T cells display hard-to-detect attraction to Plasmodium- infected hepatocytes. Front Bioinf 1:1–15. https://doi.org/10.3389/fbinf.2021.770448
Acknowledgements
This work was supported by the NIH (R01GM118553 and R01AI158963) awards to VVG.
Author information
Authors and Affiliations
Contributions
Mathematical analyses and analytical proofs were done by H.R. Simulations of cluster formations were done primarily by V.V.G. H.R. wrote the first draft of the paper with all authors contributing to the final draft.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rajakaruna, H., Ganusov, V.V. Mathematical Modeling to Guide Experimental Design: T Cell Clustering as a Case Study. Bull Math Biol 84, 103 (2022). https://doi.org/10.1007/s11538-022-01063-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-022-01063-x