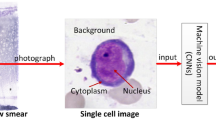
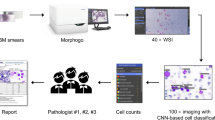
Abstract
Standardized morphological evaluation in pathology is usually qualitative. Classifying and qualitatively analyzing the nucleated cells in the bone marrow aspirate images based on morphology is crucial for the diagnosis of acute myoid leukemia (AML), acute lymphoblastic leukemia (ALL), and Myelodysplastic syndrome (MDS), etc. However, it is time-consuming and difficult to accurately identify nucleated cells and calculate the percentage of the cells because of the complexity of bone marrow aspirate images. This paper proposed a deep learning analysis model of bone marrow aspirate images, termed Cell Detection and Confirmation Network (CDC-NET), for the aided diagnosis of AML by improving the accuracy of cell detection and recognition. Specifically, we take the nucleated cells in the bone marrow aspirate images as the detection objects to establish the model. Since some cells from different categories have similar morphology, classification error is inevitable. We design a confirmation network in which multiple trained classifiers work as pathologists to confirm the cell category by a voting method. To demonstrate the effectiveness of the proposed approach, experiments on clinical microscopic datasets are conducted. The Recall and Precision of CDC-NET are 78.54% and 91.74% respectively, and the missed rate of our method is lower than those of the other popular methods. The experimental results demonstrated that the proposed model has the potential for the pathological analysis of aspirate smears and the aided diagnosis of AML.
Graphical abstract











Similar content being viewed by others
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Bruneau J, Molina TJ (2020) WHO Classification of tumors of hematopoietic and lymphoid tissues. In: Molina TJ (eds) Hematopathology. Encyclopedia of Pathology. Springer, Cham, pp 501–505. https://doi.org/10.1007/978-3-319-95309-0_3817
Bonnet D, Dick JE (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3(7):730–737
Porcu S, Loddo A, Putzu L, Di Ruberto C (2018) White blood cells counting via vector field convolution nuclei segmentation. In: Proceedings of the 13th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (4):227–234
Das DK, Maiti AK, Chakraborty C (2018) Automated identification of normoblast cell from human peripheral blood smear images. J Microsc 269(3):310–320
Ghribi O, Maalej A, Sellami L, Slima MB, Maalej MA, Mahfoudh KB, Dammak M, Mhiri C, Hamida AB (2019) Advanced methodology for multiple sclerosis lesion exploring: Towards a computer aided diagnosis system. Biomed Signal Process Control 49:274–288
Matek C, Schwarz S, Spiekermann K, Marr C (2019) Human-level recognition of blast cells in acute myeloid leukaemia with convolutional neural networks. Nat Mach Intell 1(11):538–544
El Alaoui Y, Elomri A, Qaraqe M, Padmanabhan R, YasinTaha R, El Omri H, El Omri A, Aboumarzouk O (2022) A review of artificial intelligence applications in hematology management: current practices and future prospects. J Med Internet Res 24(7):e36490
Raina R, Gondhi NK, Chaahat R, Singh D, Kaur M, Lee HN (2023) A systematic review on acute leukemia detection using deep learning techniques. Arch Comput Methods Eng 30(1):251–270
Saleem S, Amin J, Sharif M, Mallah GA, Kadry S, Gandomi AH (2022) Leukemia segmentation and classification: A comprehensive survey. Comput Biol Med 150:106028
Akram N, Adnan S, Asif M, Imran SMA, Yasir MN, Naqvi RA, Hussain D (2022) Exploiting the multiscale information fusion capabilities for aiding the leukemia diagnosis through white blood cells segmentation. IEEE Access 10:48747–48760
Mustaqim T, Fatichah C, Suciati N (2023) Deep learning for the detection of acute lymphoblastic leukemia subtypes on microscopic images: a systematic literature review. IEEE Access (11):16108–16127. https://doi.org/10.1109/ACCESS.2023.3245128
Li N, Fan L, Xu H, Zhang X, Bai Z, Li M, Xiong S, Jiang L, Yang J, Chen S, Qiao Y (2023) An AI-Aided diagnostic framework for hematologic neoplasms based on morphologic features and medical expertise. Lab Invest 103(4):100055
Goutam D, Sailaja S (2015) Classification of acute myelogenous leukemia in blood microscopic images using supervised classifier. In: 2015 IEEE International Conference on Engineering and Technology (ICETECH). IEEE, pp 1–5
Li Y, Zhu R, Mi L, Cao Y, Yao D (2016) Segmentation of white blood cell from acute lymphoblastic leukemia images using dual-threshold method. Comput Math Methods Med 2016:9514707
Aris TA, Nasir AA, Mustafa WA (2018) Analysis of distance transforms for watershed segmentation on chronic leukaemia images. J Telecommun Electron Comput Eng (JTEC) 10(1–16):51–56
Song TH, Sanchez V, Daly HE, Rajpoot NM (2018) Simultaneous cell detection and classification in bone marrow histology images. IEEE J Biomed Health Inform 23(4):1469–1476
Yang S, Liu X, Zheng Z, Wang W, Ma X (2021) Fusing medical image features and clinical features with deep learning for computer-aided diagnosis. arXiv preprint arXiv:2103.05855, 2021
Fan H, Zhang F, Xi L, Li Z, Liu G, Xu Y (2019) LeukocyteMask: An automated localization and segmentation method for leukocyte in blood smear images using deep neural networks. J Biophotonics 12(7):e201800488
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Ioffe S, Szegedy C (2015) Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International conference on machine learning, pp 448–456. pmlr
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Yu W, Chang J, Yang C, Zhang L, Shen H, Xia Y, Sha J (2017) Automatic classification of leukocytes using deep neural network. In: 2017 IEEE 12th international conference on ASIC (ASICON). IEEE, pp 1041–1044
Suriya M, Chandran V, Sumithra MG (2022) Enhanced deep convolutional neural network for malarial parasite classification. Int J Comput Appl 44(12):1113–1122
Zerouaoui H, Idri A, Nakach FZ, Hadri RE (2021) Breast fine needle cytological classification using deep hybrid architectures. In: Computational Science and Its Applications–ICCSA 2021: 21st International Conference, Cagliari, Italy, September 13–16, 2021, Proceedings, Part II 21. Springer International Publishing, pp 186–202
Ren S, He K, Girshick R, Sun J (2017) Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 39(06):1137–1149
Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu CY, Berg AC (2016) Ssd: Single shot multibox detector. In: Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part I 14. Springer International Publishing, pp 21–37
Redmon J, Divvala S, Girshick R, Farhadi A (2016) You only look once: Unified, real-time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 779–788
Redmon J, Farhadi A (2017) YOLO9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7263–7271
Redmon J, Farhadi A (2018) YOLOv3: an incremental improvement. https://doi.org/10.48550/arXiv.1804.02767
Bochkovskiy A, Wang CY, Liao HYM (2020) Yolov4: optimal speed and accuracy of object detection. https://doi.org/10.48550/arXiv.2004.10934
Su J, Han J, Song J (2021) A benchmark bone marrow aspirate smear dataset and a multi-scale cell detection model for the diagnosis of hematological disorders. Comput Med Imaging Graph 90:101912
Rahman, MA, Wang, Y (2016) Optimizing intersection-over-union in deep neural networks for image segmentation. In: Bebis G, et al. Advances in Visual Computing. ISVC 2016. Lecture Notes in Computer Science, vol 10072. Springer, Cham. https://doi.org/10.1007/978-3-319-50835-1_22
Raghavan V, Bollmann P, Jung GS (1989) A critical investigation of recall and precision as measures of retrieval system performance. ACM Trans Inf Syst (TOIS) 7(3):205–229
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (No. 52001039), National Natural Science Foundation of China under Grand (No.52171310), Shandong Natural Science Foundation in China (No. ZR2019LZH005), Research fund from Science and Technology on Underwater Vehicle Technology Laboratory (No.2021JCJQ-SYSJJ-LB06903). University Innovation Team Project of Jinan (No.2019GXRC015). Science and technology improvement project for small and medium-sized enterprises in Shandong Province (No. 2021TSGC1012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• A Cell Detection and Confirmation Network is designed to achieve automatic analysis of the bone marrow aspirate images to assist in AML diagnosis.
• It simulates pathologists by introducing a voting mechanism to balance their perspectives and further achieve consistent results, and it possesses excellent scalability.
• The experimental results demonstrated that the proposed model has the potential for the pathological analysis of aspirate smears and the aided diagnosis of AML.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Su, J., Liu, Y., Zhang, J. et al. CDC-NET: a cell detection and confirmation network of bone marrow aspirate images for the aided diagnosis of AML. Med Biol Eng Comput 62, 575–589 (2024). https://doi.org/10.1007/s11517-023-02955-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02955-3