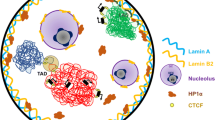
Abstract
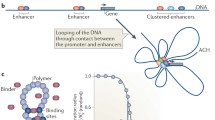
Eukaryotic genomes are densely packaged into hierarchical three-dimensional (3D) structures that contain information about gene regulation and many other biological processes. With the development of imaging and sequencing-based technologies, 3D genome studies have revealed that the high-order chromatin structure is composed of hierarchical levels, including chromosome territories, A/B compartments, topologically associated domains, and chromatin loops. However, how this chromatin architecture is formed and maintained is not completely clear. In this review, we introduce experimental methods to investigate the 3D genome, review major architectural proteins that regulate 3D chromatin organization in mammalian cells, such as CTCF (CCCTC-binding factor), cohesin, lamins, and transcription factors, and discuss relevant mechanisms such as phase separation.
Similar content being viewed by others
References
Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M.A., and Nasmyth, K. (2001). Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472.
Alipour, E., and Marko, J.F. (2012). Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res 40, 11202–11212.
Apostolou, E., and Thanos, D. (2008). Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-α gene expression. Cell 134, 85–96.
Arumugam, P., Gruber, S., Tanaka, K., Haering, C.H., Mechtler, K., and Nasmyth, K. (2003). ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol 13, 1941–1953.
Bantignies, F., Roure, V., Comet, I., Leblanc, B., Schuettengruber, B., Bonnet, J., Tixier, V., Mas, A., and Cavalli, G. (2011). Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144, 214–226.
Bao, L., Zhou, M., and Cui, Y. (2008). CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res 36, D83–D87.
Beagan, J.A., Gilgenast, T.G., Kim, J., Plona, Z., Norton, H.K., Hu, G., Hsu, S.C., Shields, E.J., Lyu, X., Apostolou, E., et al. (2016). Local genome topology can exhibit an incompletely rewired 3D-folding state during somatic cell reprogramming. Cell Stem Cell 18, 611–624.
Beagrie, R.A., Scialdone, A., Schueler, M., Kraemer, D.C.A., Chotalia, M., Xie, S.Q., Barbieri, M., de Santiago, I., Lavitas, L.M., Branco, M.R., et al. (2017). Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524.
Boija, A., Klein, I.A., Sabari, B.R., Dall’Agnese, A., Coffey, E.L., Zamudio, A.V., Li, C.H., Shrinivas, K., Manteiga, J.C., Hannett, N. M., et al. (2018). Transcription factors activate genes through the phaseseparation capacity of their activation domains. Cell 175, 1842–1855.e16.
Boulay, G., Sandoval, G.J., Riggi, N., Iyer, S., Buisson, R., Naigles, B., Awad, M.E., Rengarajan, S., Volorio, A., McBride, M.J., et al. (2017). Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178.e19.
Bronshtein, I., Kepten, E., Kanter, I., Berezin, S., Lindner, M., Redwood, A.B., Mai, S., Gonzalo, S., Foisner, R., Shav-Tal, Y., et al. (2015). Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat Commun 6, 8044.
Chang, L., Li, M., Shao, S., Xue, B., Hou, Y., Zhang, Y., Li, R., Li, C., and Sun, Y. (2019). Chromatin-lamin B1 interaction promotes genomic compartmentalization and constrains chromatin dynamics. bioRxiv, 601849.
Chen, H., Tian, Y., Shu, W., Bo, X., and Wang, S. (2012). Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS ONE 7, e41374.
Cho, W.K., Spille, J.H., Hecht, M., Lee, C., Li, C., Grube, V., and Cisse, I.I. (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415.
Chong, S., Dugast-Darzacq, C., Liu, Z., Dong, P., Dailey, G.M., Cattoglio, C., Heckert, A., Banala, S., Lavis, L., Darzacq, X., et al. (2018). Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555.
Chowdhary, S., Kainth, A.S., Pincus, D., and Gross, D.S. (2019). Heat shock factor 1 drives intergenic association of its target gene loci upon heat shock. Cell Rep 26, 18–28.e5.
Colognori, D., Sunwoo, H., Kriz, A.J., Wang, C.Y., and Lee, J.T. (2019). Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol Cell 74, 101–117.e10.
Corces, M.R., Buenrostro, J.D., Wu, B., Greenside, P.G., Chan, S.M., Koenig, J.L., Snyder, M.P., Pritchard, J.K., Kundaje, A., Greenleaf, W. J., et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48, 1193–1203.
Cruz-Molina, S., Respuela, P., Tebartz, C., Kolovos, P., Nikolic, M., Fueyo, R., van Ijcken, W.F.J., Grosveld, F., Frommolt, P., Bazzi, H., et al. (2017). PRC2 facilitates the regulatory topology required for poised enhancer function during pluripotent stem cell differentiation. Cell Stem Cell 20, 689–705.e9.
Dall’Agnese, A., Caputo, L., Nicoletti, C., di Iulio, J., Schmitt, A., Gatto, S., Diao, Y., Ye, Z., Forcato, M., Perera, R., et al. (2019). Transcription factor-directed re-wiring of chromatin architecture for somatic cell nuclear reprogramming toward trans-differentiation. Mol Cell 76, 453–472.e8.
Dekker, J., Belmont, A.S., Guttman, M., Leshyk, V.O., Lis, J.T., Lomvardas, S., Mirny, L.A., O’Shea, C.C., Park, P.J., Ren, B., et al. (2017). The 4D nucleome project. Nature 549, 219–226.
Dekker, J., and Mirny, L. (2016). The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121.
Dekker, J., Rippe, K., Dekker, M., and Kleckner, N. (2002). Capturing chromosome conformation. Science 295, 1306–1311.
Deng, W., Lee, J., Wang, H., Miller, J., Reik, A., Gregory, P.D., Dean, A., and Blobel, G.A. (2012). Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244.
Denisenko, O., and Bomsztyk, K. (2002). Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length. Mol Cell Biol 22, 286–297.
Dixon, J.R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., Hu, M., Liu, J. S., and Ren, B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380.
Dolgin, E. (2017). DNA’s secret weapon against knots and tangles. Nature 544, 284–286.
Falk, M., Feodorova, Y., Naumova, N., Imakaev, M., Lajoie, B.R., Leonhardt, H., Joffe, B., Dekker, J., Fudenberg, G., Solovei, I., et al. (2019). Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570, 395–399.
Fan, H., Lv, P., Huo, X., Wu, J., Wang, Q., Cheng, L., Liu, Y., Tang, Q.Q., Zhang, L., Zhang, F., et al. (2018). The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res 28, 192–202.
Fang, R., Yu, M., Li, G., Chee, S., Liu, T., Schmitt, A.D., and Ren, B. (2016). Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res 26, 1345–1348.
Feng, Z., Chen, X., Wu, X., and Zhang, M. (2019). Formation of biological condensates via phase separation: Characteristics, analytical methods, and physiological implications. J Biol Chem 294, 14823–14835.
Filippova, G.N., Fagerlie, S., Klenova, E.M., Myers, C., Dehner, Y., Goodwin, G., Neiman, P.E., Collins, S.J., and Lobanenkov, V.V. (1996). An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16, 2802–2813.
Flavahan, W.A., Drier, Y., Liau, B.B., Gillespie, S.M., Venteicher, A.S., Stemmer-Rachamimov, A.O., Suvà, M.L., and Bernstein, B.E. (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114.
Flyamer, I.M., Gassler, J., Imakaev, M., Brandão, H.B., Ulianov, S.V., Abdennur, N., Razin, S.V., Mirny, L.A., and Tachibana-Konwalski, K. (2017). Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114.
Fudenberg, G., Imakaev, M., Lu, C., Goloborodko, A., Abdennur, N., and Mirny, L.A. (2016). Formation of chromosomal domains by loop extrusion. Cell Rep 15, 2038–2049.
Fullwood, M.J., Liu, M.H., Pan, Y.F., Liu, J., Xu, H., Mohamed, Y.B., Orlov, Y.L., Velkov, S., Ho, A., Mei, P.H., et al. (2009). An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64.
Gao, P., Xia, J.H., Sipeky, C., Dong, X.M., Zhang, Q., Yang, Y., Zhang, P., Cruz, S.P., Zhang, K., Zhu, J., et al. (2018). Biology and clinical implications of the 19q13 aggressive prostate cancer susceptibility locus. Cell 174, 576–589.e18.
Garee, J.P., and Oesterreich, S. (2010). SAFB1’s multiple functions in biological control-lots still to be done! J Cell Biochem 109, 312–319.
Gassler, J., Brandão, H.B., Imakaev, M., Flyamer, I.M., Ladstätter, S., Bickmore, W.A., Peters, J.M., Mirny, L.A., and Tachibana, K. (2017). A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36, 3600–3618.
Gatewood, J.M., Cook, G.R., Balhorn, R., Schmid, C.W., and Bradbury, E. M. (1990). Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem 265, 20662–20666.
Geeven, G., Zhu, Y., Kim, B.J., Bartholdy, B.A., Yang, S.M., Macfarlan, T. S., Gifford, W.D., Pfaff, S.L., Verstegen, M.J.A.M., Pinto, H., et al. (2015). Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol 16, 289.
Gesson, K., Rescheneder, P., Skoruppa, M.P., von Haeseler, A., Dechat, T., and Foisner, R. (2016). A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res 26, 462–473.
Getzenberg, R.H., Pienta, K.J., Ward, W.S., and Coffey, D.S. (1991). Nuclear structure and the three-dimensional organization of DNA. J Cell Biochem 47, 289–299.
Gibson, B.A., Doolittle, L.K., Schneider, M.W.G., Jensen, L.E., Gamarra, N., Henry, L., Gerlich, D.W., Redding, S., and Rosen, M.K. (2019). Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21.
Giorgetti, L., and Heard, E. (2016). Closing the loop: 3C versus DNA FISH. Genome Biol 17, 215.
Gruenbaum, Y., and Foisner, R. (2015). Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 84, 131–164.
Gu, Z., Jin, K., Crabbe, M.J.C., Zhang, Y., Liu, X., Huang, Y., Hua, M., Nan, P., Zhang, Z., and Zhong, Y. (2016). Enrichment analysis of Alu elements with different spatial chromatin proximity in the human genome. Protein Cell 7, 250–266.
Guelen, L., Pagie, L., Brasset, E., Meuleman, W., Faza, M.B., Talhout, W., Eussen, B.H., de Klein, A., Wessels, L., de Laat, W., et al. (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951.
Guo, Y., Xu, Q., Canzio, D., Shou, J., Li, J., Gorkin, D.U., Jung, I., Wu, H., Zhai, Y., Tang, Y., et al. (2015). CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910.
Haarhuis, J.H.I., van der Weide, R.H., Blomen, V.A., Yáñez-Cuna, J.O., Amendola, M., van Ruiten, M.S., Krijger, P.H.L., Teunissen, H., Medema, R.H., van Steensel, B., et al. (2017). The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707.e14.
Hansen, A.S., Hsieh, T.H.S., Cattoglio, C., Pustova, I., Saldaña-Meyer, R., Reinberg, D., Darzacq, X., and Tjian, R. (2019). Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol Cell 76, 395–411.e13.
Hao, N., Shearwin, K.E., and Dodd, I.B. (2019). Positive and negative control of enhancer-promoter interactions by other DNA loops generates specificity and tunability. Cell Rep 26, 2419–2433.e3.
Hark, A.T., Schoenherr, C.J., Katz, D.J., Ingram, R.S., Levorse, J.M., and Tilghman, S.M. (2000). CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489.
Hauf, S., Waizenegger, I.C., and Peters, J.M. (2001). Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320–1323.
Hnisz, D., Day, D.S., and Young, R.A. (2016). Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167, 1188–1200.
Hnisz, D., Shrinivas, K., Young, R.A., Chakraborty, A.K., and Sharp, P.A. (2017). A phase separation model for transcriptional control. Cell 169, 13–23.
Hou, Y., Guo, H., Cao, C., Li, X., Hu, B., Zhu, P., Wu, X., Wen, L., Tang, F., Huang, Y., et al. (2016). Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 26, 304–319.
Hsieh, T.H.S., Weiner, A., Lajoie, B., Dekker, J., Friedman, N., and Rando, O.J. (2015). Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell 162, 108–119.
Hu, B., Wang, N., Bi, X., Karaaslan, E.S., Weber, A.L., Zhu, W., Berendzen, K.W., and Liu, C. (2019). Plant lamin-like proteins mediate chromatin tethering at the nuclear periphery. Genome Biol 20, 87.
Hug, C.B., Grimaldi, A.G., Kruse, K., and Vaquerizas, J.M. (2017). Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169, 216–228.e19.
Huo, X., Ji, L., Zhang, Y., Lv, P., Cao, X., Wang, Q., Yan, Z., Dong, S., Du, D., Zhang, F., et al. (2020). The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol Cell 77, 368–383.e7.
Hyman, A.A., Weber, C.A., and Jülicher, F. (2014). Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 30, 39–58.
Jäger, R., Migliorini, G., Henrion, M., Kandaswamy, R., Speedy, H.E., Heindl, A., Whiffin, N., Carnicer, M.J., Broome, L., Dryden, N., et al. (2015). Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun 6, 6178.
Kaaij, L.J.T., Mohn, F., van der Weide, R.H., de Wit, E., and Bühler, M. (2019). The ChAHP complex counteracts chromatin looping at CTCF sites that emerged from SINE expansions in mouse. Cell 178, 1437–1451.e14.
Khanna, N., Hu, Y., and Belmont, A.S. (2014). HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol 24, 1138–1144.
Kim, T.H., Abdullaev, Z.K., Smith, A.D., Ching, K.A., Loukinov, D.I., Green, R.D., Zhang, M.Q., Lobanenkov, V.V., and Ren, B. (2007). Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245.
Kirby, T.J., and Lammerding, J. (2018). Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol 20, 373–381.
Kojic, A., Cuadrado, A., De Koninck, M., Giménez-Llorente, D., Rodríguez-Corsino, M., Gómez-López, G., Le Dily, F., Marti-Renom, M.A., and Losada, A. (2018). Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol 25, 496–504.
Lafont, A.L., Song, J., and Rankin, S. (2010). Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci USA 107, 20364–20369.
Langer-Safer, P.R., Levine, M., and Ward, D.C. (1982). Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci USA 79, 4381–4385.
Larson, A.G., Elnatan, D., Keenen, M.M., Trnka, M.J., Johnston, J.B., Burlingame, A.L., Agard, D.A., Redding, S., and Narlikar, G.J. (2017). Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240.
Levasseur, D.N., Wang, J., Dorschner, M.O., Stamatoyannopoulos, J.A., and Orkin, S.H. (2008). Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev 22, 575–580.
Li, T., Jia, L., Cao, Y., Chen, Q., and Li, C. (2018). OCEAN-C: mapping hubs of open chromatin interactions across the genome reveals gene regulatory networks. Genome Biol 19, 54.
Liang, Z., Li, G., Wang, Z., Djekidel, M.N., Li, Y., Qian, M.P., Zhang, M. Q., and Chen, Y. (2017). BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nat Commun 8, 1622.
Lieberman-Aiden, E., van Berkum, N.L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., Amit, I., Lajoie, B.R., Sabo, P.J., Dorschner, M.O., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293.
Lin, D., Hong, P., Zhang, S., Xu, W., Jamal, M., Yan, K., Lei, Y., Li, L., Ruan, Y., Fu, Z.F., et al. (2018). Digestion-ligation-only Hi-C is an efficient and cost-effective method for chromosome conformation capture. Nat Genet 50, 754–763.
Linnemann, A.K., and Krawetz, S.A. (2009). Maintenance of a functional higher order chromatin structure: The role of the nuclear matrix in normal and disease states. Gene Ther Mol Biol 13, 231–243.
Lobanenkov, V.V., Nicolas, R.H., Adler, V.V., Paterson, H., Klenova, E.M., Polotskaja, A.V., and Goodwin, G.H. (1990). A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5, 1743–1753.
Lomvardas, S., Barnea, G., Pisapia, D.J., Mendelsohn, M., Kirkland, J., and Axel, R. (2006). Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413.
Losada, A., Hirano, M., and Hirano, T. (1998). Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12, 1986–1997.
Lu, H., Yu, D., Hansen, A.S., Ganguly, S., Liu, R., Heckert, A., Darzacq, X., and Zhou, Q. (2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323.
Lu, J.Y., Chang, L., Li, T., Wang, T., Yin, Y., Zhan, G., Zhang, K., Percharde, M., Wang, L., Peng, Q., et al. (2019). L1 and B1 repeats blueprint the spatial organization of chromatin. bioRxiv, 802173.
Lupiáñez, D.G., Kraft, K., Heinrich, V., Krawitz, P., Brancati, F., Klopocki, E., Horn, D., Kayserili, H., Opitz, J.M., Laxova, R., et al. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025.
Ma, W., Ay, F., Lee, C., Gulsoy, G., Deng, X., Cook, S., Hesson, J., Cavanaugh, C., Ware, C.B., Krumm, A., et al. (2015). Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods 12, 71–78.
Maass, P.G., Barutcu, A.R., Weiner, C.L., and Rinn, J.L. (2018). Inter-chromosomal contact properties in live-cell imaging and in Hi-C. Mol Cell 69, 1039–1045.e3.
Maeshima, K., Ide, S., Hibino, K., and Sasai, M. (2016). Liquid-like behavior of chromatin. Curr Opin Genet Dev 37, 36–45.
Mao, Y.S., Zhang, B., and Spector, D.L. (2011). Biogenesis and function of nuclear bodies. Trends Genet 27, 295–306.
Markenscoff-Papadimitriou, E., Allen, W.E., Colquitt, B.M., Goh, T., Murphy, K.K., Monahan, K., Mosley, C.P., Ahituv, N., and Lomvardas, S. (2014). Enhancer interaction networks as a means for singular olfactory receptor expression. Cell 159, 543–557.
Maurer, M., and Lammerding, J. (2019). The driving force: nuclear mechanotransduction in cellular function, fate, and disease. Annu Rev Biomed Eng 21, 443–468.
Meaburn, K.J., and Misteli, T. (2007). Chromosome territories. Nature 445, 379–381.
Meistrich, M.L., Mohapatra, B., Shirley, C.R., and Zhao, M. (2003). Roles of transition nuclear proteins in spermiogenesis. Chromosoma 111, 483–488.
Meuleman, W., Peric-Hupkes, D., Kind, J., Beaudry, J.B., Pagie, L., Kellis, M., Reinders, M., Wessels, L., and van Steensel, B. (2013). Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23, 270–280.
Mifsud, B., Tavares-Cadete, F., Young, A.N., Sugar, R., Schoenfelder, S., Ferreira, L., Wingett, S.W., Andrews, S., Grey, W., Ewels, P.A., et al. (2015). Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47, 598–606.
Monahan, K., Horta, A., and Lomvardas, S. (2019). LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565, 448–453.
Mumbach, M.R., Rubin, A.J., Flynn, R.A., Dai, C., Khavari, P.A., Greenleaf, W.J., and Chang, H.Y. (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods 13, 919–922.
Nagano, T., Lubling, Y., Várnai, C., Dudley, C., Leung, W., Baran, Y., Mendelson Cohen, N., Wingett, S., Fraser, P., and Tanay, A. (2017). Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67.
Nasmyth, K. (2001). Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35, 673–745.
Nasmyth, K., and Haering, C.H. (2009). Cohesin: its roles and mechanisms. Annu Rev Genet 43, 525–558.
Naumova, N., Imakaev, M., Fudenberg, G., Zhan, Y., Lajoie, B.R., Mirny, L.A., and Dekker, J. (2013). Organization of the mitotic chromosome. Science 342, 948–953.
Nikitina, T., Shi, X., Ghosh, R.P., Horowitz-Scherer, R.A., Hansen, J.C., and Woodcock, C.L. (2007). Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol 27, 864–877.
Niu, L., Shen, W., Huang, Y., He, N., Zhang, Y., Sun, J., Wan, J., Jiang, D., Yang, M., Tse, Y.C., et al. (2019). Amplification-free library preparation with SAFE Hi-C uses ligation products for deep sequencing to improve traditional Hi-C analysis. Commun Biol 2, 267.
Nora, E.P., Goloborodko, A., Valton, A.L., Gibcus, J.H., Uebersohn, A., Abdennur, N., Dekker, J., Mirny, L.A., and Bruneau, B.G. (2017). Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22.
Nora, E.P., Lajoie, B.R., Schulz, E.G., Giorgetti, L., Okamoto, I., Servant, N., Piolot, T., van Berkum, N.L., Meisig, J., Sedat, J., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385.
Patel, B., Kang, Y., Cui, K., Litt, M., Riberio, M.S.J., Deng, C., Salz, T., Casada, S., Fu, X., Qiu, Y., et al. (2014). Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia 28, 349–361.
Pederson, T. (2000). Half a century of “the nuclear matrix”. Mol Biol Cell 11, 799–805.
Pederson, T. (2011). The nucleolus. Cold Spring Harb Perspect Biol 3, a000638.
Peric-Hupkes, D., Meuleman, W., Pagie, L., Bruggeman, S.W.M., Solovei, I., Brugman, W., Gräf, S., Flicek, P., Kerkhoven, R.M., van Lohuizen, M., et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38, 603–613.
Qamar, S., Wang, G.Z., Randle, S.J., Ruggeri, F.S., Varela, J.A., Lin, J.Q., Phillips, E.C., Miyashita, A., Williams, D., Ströhl, F., et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15.
Quinodoz, S.A., Ollikainen, N., Tabak, B., Palla, A., Schmidt, J.M., Detmar, E., Lai, M.M., Shishkin, A.A., Bhat, P., Takei, Y., et al. (2018). Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e24.
Ramani, V., Deng, X., Qiu, R., Gunderson, K.L., Steemers, F.J., Disteche, C.M., Noble, W.S., Duan, Z., and Shendure, J. (2017). Massively multiplex single-cell Hi-C. Nat Methods 14, 263–266.
Rao, S.S.P., Huntley, M.H., Durand, N.C., Stamenova, E.K., Bochkov, I.D., Robinson, J.T., Sanborn, A.L., Machol, I., Omer, A.D., Lander, E.S., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680.
Rao, S.S.P., Huang, S.C., Glenn St Hilaire, B., Engreitz, J.M., Perez, E.M., Kieffer-Kwon, K.R., Sanborn, A.L., Johnstone, S.E., Bascom, G.D., Bochkov, I.D., et al. (2017). Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24.
Sabari, B.R., Dall’Agnese, A., Boija, A., Klein, I.A., Coffey, E.L., Shrinivas, K., Abraham, B.J., Hannett, N.M., Zamudio, A.V., Manteiga, J.C., et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958.
Saldaña-Meyer, R., Rodriguez-Hernaez, J., Escobar, T., Nishana, M., Jácome-López, K., Nora, E.P., Bruneau, B.G., Tsirigos, A., Furlan-Magaril, M., Skok, J., et al. (2019). RNA interactions are essential for CTCF-mediated genome organization. Mol Cell 76, 412–422.e5.
Sanyal, A., Lajoie, B.R., Jain, G., and Dekker, J. (2012). The long-range interaction landscape of gene promoters. Nature 489, 109–113.
Schoenfelder, S., Sugar, R., Dimond, A., Javierre, B.M., Armstrong, H., Mifsud, B., Dimitrova, E., Matheson, L., Tavares-Cadete, F., Furlan-Magaril, M., et al. (2015). Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet 47, 1179–1186.
Sequeira-Mendes, J., and Gutierrez, C. (2016). Genome architecture: from linear organisation of chromatin to the 3D assembly in the nucleus. Chromosoma 125, 455–469.
Shao, S., Chang, L., Sun, Y., Hou, Y., Fan, X., and Sun, Y. (2018). Multiplexed sgRNA expression allows versatile single nonrepetitive DNA labeling and endogenous gene regulation. ACS Synth Biol 7, 176–186.
Shao, S., Zhang, W., Hu, H., Xue, B., Qin, J., Sun, C., Sun, Y., Wei, W., and Sun, Y. (2016). Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res 44, e86.
Shin, Y., Chang, Y.C., Lee, D.S.W., Berry, J., Sanders, D.W., Ronceray, P., Wingreen, N.S., Haataja, M., and Brangwynne, C.P. (2019). Liquid nuclear condensates mechanically sense and restructure the genome. Cell 176, 1518.
Solovei, I., Wang, A.S., Thanisch, K., Schmidt, C.S., Krebs, S., Zwerger, M., Cohen, T.V., Devys, D., Foisner, R., Peichl, L., et al. (2013). LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598.
Stadhouders, R., Vidal, E., Serra, F., Di Stefano, B., Le Dily, F., Quilez, J., Gomez, A., Collombet, S., Berenguer, C., Cuartero, Y., et al. (2018). Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet 50, 238–249.
Stevens, T.J., Lando, D., Basu, S., Atkinson, L.P., Cao, Y., Lee, S.F., Leeb, M., Wohlfahrt, K.J., Boucher, W., O’Shaughnessy-Kirwan, A., et al. (2017). 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64.
Strom, A.R., Emelyanov, A.V., Mir, M., Fyodorov, D.V., Darzacq, X., and Karpen, G.H. (2017). Phase separation drives heterochromatin domain formation. Nature 547, 241–245.
Sumara, I., Vorlaufer, E., Gieffers, C., Peters, B.H., and Peters, J.M. (2000). Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151, 749–762.
Tan, L., Xing, D., Chang, C.H., Li, H., and Xie, X.S. (2018). Three-dimensional genome structures of single diploid human cells. Science 361, 924–928.
Tang, Z., Luo, O.J., Li, X., Zheng, M., Zhu, J.J., Szalaj, P., Trzaskoma, P., Magalska, A., Wlodarczyk, J., Ruszczycki, B., et al. (2015). CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163, 1611–1627.
Tatavosian, R., Kent, S., Brown, K., Yao, T., Duc, H.N., Huynh, T.N., Zhen, C.Y., Ma, B., Wang, H., and Ren, X. (2019). Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J Biol Chem 294, 1451–1463.
Tedeschi, A., Wutz, G., Huet, S., Jaritz, M., Wuensche, A., Schirghuber, E., Davidson, I.F., Tang, W., Cisneros, D.A., Bhaskara, V., et al. (2013). Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501, 564–568.
Thakur, J., Fang, H., Llagas, T., Disteche, C.M., and Henikoff, S. (2019). Architectural RNA is required for heterochromatin organization. bioRxiv, 784835.
Toth, A., Ciosk, R., Uhlmann, F., Galova, M., Schleiffer, A., and Nasmyth, K. (1999). Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13, 320–333.
Ulianov, S.V., Doronin, S.A., Khrameeva, E.E., Kos, P.I., Luzhin, A.V., Starikov, S.S., Galitsyna, A.A., Nenasheva, V.V., Ilyin, A.A., Flyamer, I.M., et al. (2019). Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat Commun 10, 1176.
Vietri Rudan, M., Barrington, C., Henderson, S., Ernst, C., Odom, D.T., Tanay, A., and Hadjur, S. (2015). Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 10, 1297–1309.
Wang, L., Gao, Y., Zheng, X., Liu, C., Dong, S., Li, R., Zhang, G., Wei, Y., Qu, H., Li, Y., et al. (2019). Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell 76, 646–659.e6.
Wang, Q., Sawyer, I.A., Sung, M.H., Sturgill, D., Shevtsov, S.P., Pegoraro, G., Hakim, O., Baek, S., Hager, G.L., and Dundr, M. (2016a). Cajal bodies are linked to genome conformation. Nat Commun 7, 10966.
Wang, S., Su, J.H., Beliveau, B.J., Bintu, B., Moffitt, J.R., Wu, C., and Zhuang, X. (2016b). Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602.
Wei, Z., Gao, F., Kim, S., Yang, H., Lyu, J., An, W., Wang, K., and Lu, W. (2013). Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell 13, 36–47.
Weintraub, A.S., Li, C.H., Zamudio, A.V., Sigova, A.A., Hannett, N.M., Day, D.S., Abraham, B.J., Cohen, M.A., Nabet, B., Buckley, D.L., et al. (2017). YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588.e28.
Williamson, I., Berlivet, S., Eskeland, R., Boyle, S., Illingworth, R.S., Paquette, D., Dostie, J., and Bickmore, W.A. (2014). Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev 28, 2778–2791.
Wutz, G., Várnai, C., Nagasaka, K., Cisneros, D.A., Stocsits, R.R., Tang, W., Schoenfelder, S., Jessberger, G., Muhar, M., Hossain, M.J., et al. (2017). Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36, 3573–3599.
Yan, F., Wang, X., and Zeng, Y. (2019). 3D genomic regulation of lncRNA and Xist in X chromosome. Semin Cell Dev Biol 90, 174–180.
Yang, L., Lin, C., Jin, C., Yang, J.C., Tanasa, B., Li, W., Merkurjev, D., Ohgi, K.A., Meng, D., Zhang, J., et al. (2013). lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602.
Yoshizawa, T., Ali, R., Jiou, J., Fung, H.Y.J., Burke, K.A., Kim, S.J., Lin, Y., Peeples, W.B., Saltzberg, D., Soniat, M., et al. (2018). Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705.e22.
Zeitz, M.J., Malyavantham, K.S., Seifert, B., and Berezney, R. (2009). Matrin 3: chromosomal distribution and protein interactions. J Cell Biochem 108, 125–133.
Zheng, M., Tian, S.Z., Capurso, D., Kim, M., Maurya, R., Lee, B., Piecuch, E., Gong, L., Zhu, J.J., Li, Z., et al. (2019). Multiplex chromatin interactions with single-molecule precision. Nature 566, 558–562.
Zheng, X., Hu, J., Yue, S., Kristiani, L., Kim, M., Sauria, M., Taylor, J., Kim, Y., and Zheng, Y. (2018). Lamins organize the global three-dimensional genome from the nuclear periphery. Mol Cell 71, 802–815.e7.
Zirkel, A., Nikolic, M., Sofiadis, K., Mallm, J.P., Brackley, C.A., Gothe, H., Drechsel, O., Becker, C., Altmüller, J., Josipovic, N., et al. (2018). HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70, 730–744.e6.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) (31871266 for C.L., 21573013 and 21825401 for Y.S.), National Key Research and Development Program of China (2016YFA0100103 for C.L., 2017YFA0505302 for Y.S.), and NSFC Key Research Grant 71532001 for C.L.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance and ethics The author(s) declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Li, M., Gan, J., Sun, Y. et al. Architectural proteins for the formation and maintenance of the 3D genome. Sci. China Life Sci. 63, 795–810 (2020). https://doi.org/10.1007/s11427-019-1613-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-019-1613-3