Abstract
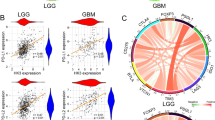
Malignant gliomas are highly heterogeneous glia-derived tumors that present an aggressive and invasive nature, with a dismal prognosis. The multi-dimensional interactions between glioma cells and other tumor microenvironment (TME) non-tumoral components constitute a challenge to finding successful treatment strategies. Several molecules, such as extracellular purines, participate in signaling events and support the immunosuppressive TME of glioma patients. The purinergic signaling and the ectoenzymes network involved in the metabolism of these extracellular nucleotides are still unexplored in the glioma TME, especially in lower-grade gliomas (LGG). Also, differences between IDH-mutant (IDH-Mut) versus wild-type (IDH-WT) gliomas are still unknown in this context. For the first time, to our knowledge, this study characterizes the TME of LGG, high-grade gliomas (HGG) IDH-Mut, and HGG IDH-WT patients regarding purinergic ectoenzymes and P1 receptors, focusing on tumor-infiltrating lymphocytes. Here, we show that ectoenzymes from both canonical and non-canonical pathways are increased in the TME when compared to the peripheral blood. We hypothesize this enhancement supports extracellular adenosine generation, hence increasing TME immunosuppression.





Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, Deimling A, Ellison DW (2021) The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 23:1231–1251. https://doi.org/10.1093/neuonc/noab106
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK (2007) Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. https://doi.org/10.1101/gad.1596707
Zou P, Xu H, Chen P, Yan Q, Zhao L, Zhao P, Gu A (2013) IDH1/IDH2 mutations define the prognosis and molecular profiles of patients with gliomas: a meta-analysis. PLoS One 8(7):e68782. https://doi.org/10.1371/journal.pone.0068782
Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH (2009) Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer 125(2):353–355. https://doi.org/10.1002/ijc.24379
Montalban-Bravo G, DiNardo CD (2018) The role of IDH mutations in acute myeloid leukemia. Future Oncol 14(10):979–993. https://doi.org/10.2217/fon-2017-0523
Wood LD et al (2007) The genomic landscapes of human breast and colorectal cancers. Science 318(5853):1108–1113. https://doi.org/10.1126/science.1145720
Vuong HG, Ngo TNM, Dunn IF (2021) Prognostic importance of IDH mutations in chondrosarcoma: An individual patient data meta-analysis. Cancer Med 10(13):4415–4423. https://doi.org/10.1002/cam4.4019
Ghiam AF, Cairns RA, Thoms J, Dal Pra A, Ahmed O, Meng A, Mak TW (2012) Bristow RG (2012) IDH mutation status in prostate cancer. Oncogene 31(33):3826. https://doi.org/10.1038/onc.2011.546
Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N (2011) Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol 178(3):1395–1402. https://doi.org/10.1016/j.ajpath.2010.12.011
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Heiden MGV, Su SM (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462(7274):739–744. https://doi.org/10.1038/nature08617
Carbonneau M et al (2016) The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat Commun 7:12700. https://doi.org/10.1038/ncomms12700
Houillier C, Wang X, Mokhtari K, Guillevin R, Laffaire J, Paris S, Boisselier B, Idbaih A, Laigle-Donadey F, Hoang-Xuan K, Sanson M, Delattre JY (2010) IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75(17):1560–1566. https://doi.org/10.1212/WNL.0b013e3181f96282
Richardson LG, Choi BD, Curry WT (2019) (R)-2-hydroxyglutarate drives immune quiescence in the tumor microenvironment of IDH-mutant gliomas. Transl Cancer Res 8(2):S167–S170. https://doi.org/10.21037/tcr.2019.01.08
Schiffer D, Annovazzi L, Casalone C, Corona C, Mellai M (2019) Glioblastoma: Microenvironment and niche concept. Cancers. https://doi.org/10.3390/cancers11010005
Gargini R, Segura-Collar B, Sánchez-Gómez P (2020) Cellular Plasticity and Tumor Microenvironment in Gliomas: The Struggle to Hit a Moving Target. Cancers (Basel) 12(6). https://doi.org/10.3390/cancers12061622
De Vleeschouwer S, Bergers G (2017) Glioblastoma: to target the tumor cell or the microenvironment? Ed., Codon Publications, Chapter 16. Brisbane, Australia. https://doi.org/10.15586/codon.glioblastoma.2017.ch16.
Knocke S, Fleischmann-Mundt B, Saborowski M, Manns MP, Kuhnel F, Wirth TC, Woller N (2016) Tailored Tumor Immunogenicity Reveals Regulation of CD4 and CD8 T Cell Responses against Cancer. Cell Rep 17(9):2234–2246. https://doi.org/10.1016/j.celrep.2016.10.086
Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B (2017) Immune microenvironment of gliomas. Lab Investig 97(5):498–518. https://doi.org/10.1038/labinvest.2017.19
Figueiró F, Muller L, Funk S, Jackson EK, Battastini AMO, Whiteside TL (2016) Phenotypic and functional characteristics of CD39high human regulatory B cells (Breg). Oncoimmunology 5 https://doi.org/10.1080/2162402X.2015.1082703
Schuler PJ, Saze Z, Hong CS, Muller L, Gillespie DG, Cheng D, Harasymczuk MM, Lang S, Jackson EK, Whiteside TL (2014) Human CD4 + CD39 + regulatory T cells produce adenosine upon co-expression of surface CD73 or contact with CD73 + exosomes or CD73 + cells. Clin Exp Immunol 177(2):531–543. https://doi.org/10.1111/cei.12354
Bavaresco L, Bernardi A, Braganhol E, Cappellari AR, Rockenbach L, Farias PF, Wink MR, Delgado-Canedo A, Battastini AMO (2008) The role of ecto-5′nucleotidase/CD73 in glioma cell line proliferation. Mol Cell Biochem 319(1):61–68. https://doi.org/10.1007/s11010-008-9877-3
Horenstein AL, Chillemi A, Zaccarello G, Bruzzone S, Quarona V, Zito A, Serra S, Malavasi F (2013) A CD38/CD203A/CD73 ectoenzymatic pathway independent of CD39 drives a novel adenosinergic loop in human T lymphocytes. Oncoimmunology 2(9):1–14. https://doi.org/10.4161/onci.26246
Di Virgilio F, Adinolfi E (2017) Extracellular purines, purinergic receptors and tumor growth. Oncogene 36(3):293–303. https://doi.org/10.1038/onc.2016.206
Ohta A, Ohta A, Madasu M, Kini R, Subramanian M, Goel N, Sitkovsky M (2009) A2A adenosine receptor may allow expansion of T cells lacking effector functions in extracellular adenosine-rich microenvironments. J Immunol 183(9):5487–5493. https://doi.org/10.4049/jimmunol.0901247
Voelter W, Zech K, Arnold P, Ludwig G (1980) Determination of selected pyrimidines, purines and their metabolites in serum and urine by reversed-phase ion-pair chromatography. J Chromatogr 199:345–354. https://doi.org/10.1016/s0021-9673(01)91386-x
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, Zhu J, Haussler D (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38(6):675–678. https://doi.org/10.1038/s41587-020-0546-8
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. https://doi.org/10.1186/1471-2105-13-134
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108. https://doi.org/10.1038/nprot.2008.73
Xu S, Shao QQ, Sun JT, Yang N, Xie Q, Wang DH, Huang QB, Huang B, Wang XY, Li XG, Qu X (2013) Synergy between the ectoenzymes CD39 and CD73 contributes to adenosinergic immunosuppression in human malignant gliomas. Neuro Oncol 15(9):1160–1172. https://doi.org/10.1093/neuonc/not067
Borsellino G, Kleinewietfeld M, Mitri DD, Sternjak A, Diamantini A, Giometto R, Hopner S, Centonze D, Bernardi G, Dell'Acqua ML, Rossini PM, Battistini L, Rotzschke O, Falk K (2007) Expression of ectonucleotidase CD39 by Foxp3 Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 110(4):1225–1233. https://doi.org/10.1182/blood-2006-12-064527
Canale FP, Ramello MC, Nuñez N, Furlan CLA, Bossio SN, Serran MG, Boari JT, Castillo AD, Ledesma M, Sedlik C, Piaggio E, Gruppi A, Rodriguez EAA, Montes CL (2018) CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8(+) T Cells. Cancer Res 78(1):115–128. https://doi.org/10.1158/0008-5472.CAN-16-2684
Zhang Y, Li W, Ma K, Zhai J, Jin Y, Zhang L, Chen C (2022) Elevated CD38 expression characterizes impaired CD8+ T cell immune response in metastatic pleural effusions. Immunol Lett 245:61–68. https://doi.org/10.1016/j.imlet.2022.04.003
Neo SY et al (2020) CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. J Clin Invest 130(3):1185–1198. https://doi.org/10.1172/JCI128895
Close HJ, Stead LF, Nsengimana J, Reilly KA, Droop A, Wurdak H, Mathew RK, Corns R, Newton-Bishop J, Melcher AA, Short SC, Cook GP, Wilson EB (2020) Expression profiling of single cells and patient cohorts identifies multiple immunosuppressive pathways and an altered NK cell phenotype in glioblastoma. Clin Exp Immunol 200(1):33–44. https://doi.org/10.1111/cei.13403
Yan Y, Li W, Liu Q, Yang K (2022) Advances in Immune Microenvironment and Immunotherapy of Isocitrate Dehydrogenase Mutated Glioma. Front Immunol 13. https://doi.org/10.3389/fimmu.2022.914618
Domínguez-Pantoja M, Lopez-Herrera G, Romero-Ramirez H, Santos-Argumedo L, Chavez-Rueda AK, Hernandez-Cueto A, Flores-Munoz M, Rodriguez-Alba JC (2018) CD38 protein deficiency induces autoimmune characteristics and its activation enhances IL-10 production by regulatory B cells. Scand J Immunol 87(6):e12664. https://doi.org/10.1111/sji.12664.
Gessi S, Merighi S, Sacchetto V, Simioni C (1808) Borea PA (2011) Adenosine receptors and cancer. Biochim Biophys Acta - Biomembr 5:1400–1412. https://doi.org/10.1016/j.bbamem.2010.09.020
Ohta A, Gorelik E, Prasad SJ, Ronchese F, Lukashev D, Wong MKK, Huang X, Caldwell S, Liu K, Smith P, Chen JF, Jackson EK, Apasov S, Abrams S, Sitkovsky M (2006) A2A adenosine receptor protects tumors from antitumor T cells. Proc Natl Acad Sci U S A 103(35):13132–13137. https://doi.org/10.1073/pnas.0605251103
Inda MDM, Bonavia R, Seoane J (2014) Glioblastoma multiforme: a look inside its heterogeneous nature. Cancers (Basel) 6(1):226–239. https://doi.org/10.3390/cancers6010226
Liu Z, Meng Q, Bartek J, Poiret T, Persson O, Rane L, Rangelova E, Illies C, Peredo IH, Luo X, Rao MV, Robertson RA, Dodoo E, Maeurer M (2017) Tumor-infiltrating lymphocytes (TILs) from patients with glioma. Oncoimmunology 6(2). https://doi.org/10.1080/2162402X.2016.1252894
DeCordova S, Shastri A, Tsolaki AG, Yasmin H, Klein L, Singh SK, Kishore U (2020) Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front Immunol 11:1402. https://doi.org/10.3389/fimmu.2020.01402
Bastid J, Regairaz A, Bonnefoy N, Dejou C, Giustiniani J, Laheurte C, Cochaud S, Laprevotte E, Funck-Bretano E, Hemon P, Gros L, Bec N, Larroque C, Alberici G, Bensussan A, Eliaou JF (2015) Inhibition of CD39 enzymatic function at the surface of tumor cells alleviates their immunosuppressive activity. Cancer Immunol Res 3(3):254–265. https://doi.org/10.1158/2326-6066.CIR-14-0018
Di Virgilio F, Falzoni S, Giuliani AL, Adinolfi E (2016) P2 receptors in cancer progression and metastatic spreading. Curr Opin Pharmacol 29:17–25. https://doi.org/10.1016/j.coph.2016.05.001
Coy S, Wang S, Stopka SA, Lin JR, Yapp C, Ritch CC, Salhi L, Baker GJ, Rashid R, Baquer G, Regan M, Khadka P, Cole KA, Hwang J, Wen PY, Bandopadhayay P, Santi M, Raedt TD, Ligon KL, Agar NYR, Sorger PK, Touat M, Santagata S (2022) Single cell spatial analysis reveals the topology of immunomodulatory purinergic signaling in glioblastoma. Nat Commun 13(1):4814. https://doi.org/10.1038/s41467-022-32430-w
Seager RJ, Hajal C, Spill F, Kamm RD, Zaman MH (2017) Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg Sci Phys Oncol 3. https://doi.org/10.1088/2057-1739/aa7e86
Ungefroren H, Sebens S, Seidl D, Lehnert H, Hass R (2011) Interaction of tumor cells with the microenvironment. Cell Commun Signal 9(18). https://doi.org/10.1186/1478-811X-9-18
Azambuja JH, Ludwig N, Yerneni S, Rao A, Braganhol E, Whiteside TL (2020) Molecular profiles and immunomodulatory activities of glioblastoma-derived exosomes. Neuro-Oncology Adv. https://doi.org/10.1093/noajnl/vdaa056
Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen JF, Enjyoji K, Linden J, Oukka M, Kuchroo VK, Strom TB, Robson SC (2007) Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med 204(6):1257–1265. https://doi.org/10.1084/jem.20062512
Desland FA, Hormigo A (2020) The CNS and the Brain Tumor Microenvironment: Implications for Glioblastoma Immunotherapy. Int J Mol Sci 21(19). https://doi.org/10.3390/ijms21197358
Sampson JH, Gunn MD, Fecci PE, Ashley DM (2020) Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer 20(1):12–25. https://doi.org/10.1038/s41568-019-0224-7
Mandapathil M, Lang S, Gorelik E, Whiteside TL (2009) Isolation of functional human regulatory T cells (Treg) from the peripheral blood based on the CD39 expression. J Immunol Methods 346:55–63. https://doi.org/10.1016/j.jim.2009.05.004
Takenaka MC, Robson SC, Quintana FJ (2016) Regulation of the T Cell Response by CD39. Trends Immunol 37(7):427–439. https://doi.org/10.1016/j.it.2016.04.009
Heimberger AB, Abou-Ghazal M, Reina-Ortiz C, Yang DS, Sun W, Qiao W, Hiraoka N, Fuller GN (2008) Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin Cancer Res 14(16):5166–5172. https://doi.org/10.1158/1078-0432.CCR-08-0320
Sayour EJ, McLendon P, McLendon R, De Leon G, Reynolds R, Kresak J, Sampson JH, Mitchell DA (2015) Increased proportion of FoxP3+ regulatory T cells in tumor infiltrating lymphocytes is associated with tumor recurrence and reduced survival in patients with glioblastoma. Cancer Immunol Immunother 64(4):419–427. https://doi.org/10.1007/s00262-014-1651-7
Vivier E, Ugolini S (2015) Regulatory natural killer cells: new players in the IL-10 anti-inflammatory response. Cell Host Microbe 6(6):493–495. https://doi.org/10.1016/j.chom.2009.12.001
Yang R, Cheng S, Luo N, Gao R, Yo K, Kang B, Wang L, Zhang Q, Fang Q, Zhang L, Li C, He A, Hu X, Peng J, Ren X, Zhang Z (2019) Distinct epigenetic features of tumor-reactive CD8+ T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis. Genome Biol 21(1):2. https://doi.org/10.1186/s13059-019-1921-y
Qi Y, Xia Y, Lin Z, Qi Y, Chen Y, Zhou Q, Zhen H, Wang J, Chang Y, Bai Q, Wang Y, Zhu Y, Xu L, Chen L, Kong Y, Zhang W, Dai B, Liu L, Guo J, Xu J (2020) Tumor-infiltrating CD39(+)CD8(+) T cells determine poor prognosis and immune evasion in clear cell renal cell carcinoma patients. Cancer Immunol Immunother 69(8):1565–1576. https://doi.org/10.1007/s00262-020-02563-2
Simoni Y et al (2018) Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557(7706):575–579. https://doi.org/10.1038/s41586-018-0130-2
Soares A, Govender L, Hughes J, Mavakla W, Kock M, Bernard C, Pienaar B, Rensburg EJ, Jacobs G, Khomba G, Stone L, Abel B, Scriba TJ, Hanekon WA (2010) Novel application of Ki67 to quantify antigen-specific in vitro lymphoproliferation. J Immunol Methods 362(1):43–50. https://doi.org/10.1016/j.jim.2010.08.007
Baitsch L, Baumgaertner P, Devevre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, Rufer N, Speiser DE (2011) Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J Clin Invest 121(6):2350–2360. https://doi.org/10.1172/JCI46102
Duhen T, Duhen R, Montler R, Moses J, Moudgil T, Miranda NF, Goodall CP, Blair TC, Fox BA, McDermott JE, Chang SC, Grunkemeier G, Leidner R, Bell RB, Weinberg AD (2018) Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun 9(1):2724. https://doi.org/10.1038/s41467-018-05072-0
Workel HH, Rooji NV, Plat A, Spierings DCJ, Fehrmann RSN, Nijman HW, Bruyn M (2020) Transcriptional Activity and Stability of CD39+CD103+CD8+ T Cells in Human High-Grade Endometrial Cancer. Int J Mol Sci 21(11). https://doi.org/10.3390/ijms21113770
Liu T, Tan J, Wu M, Fan W, Wei J, Zhu B, Guo J, Wang S, Zhou P, Zhang H, Shi L, Li J (2021) High-affinity neoantigens correlate with better prognosis and trigger potent antihepatocellular carcinoma (HCC) activity by activating CD39+CD8+ T cells. Gut 70(10):1965. https://doi.org/10.1136/gutjnl-2020-322196
DiLillo DJ, Yanaba K, Tedder TF (2010) B cells are required for optimal CD4+ and CD8+ T cell tumor immunity: therapeutic B cell depletion enhances B16 melanoma growth in mice. J Immunol 184(7):4006–4016. https://doi.org/10.4049/jimmunol.0903009
Burger JA, Wiestner A (2018) Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nat Rev Cancer 18(3):148–167. https://doi.org/10.1038/nrc.2017.121
Saze Z, Schuler P, Hong C, Cheng D (2013) Adenosine production by human B cells and B cell–mediated suppression of activated T cells. Blood 122:9–19. https://doi.org/10.1182/blood-2013-02-482406.Z.S
Quarona V, Zaccarello G, Chillemi A, Brunetti E, Singh VK, Ferrero E, Funaro A, Horenstein AL, Malavasi F (2013) CD38 and CD157: a long journey from activation markers to multifunctional molecules. Cytometry B Clin Cytom 84(4):207–217. https://doi.org/10.1002/cyto.b.21092
Bahri R, Bollinger A, Bollinger T, Orinska Z, Bulfone-Paus S (2012) Ectonucleotidase CD38 demarcates regulatory, memory-like CD8+ T cells with IFN-γ-mediated suppressor activities. PLoS One 7(9):e45234. https://doi.org/10.1371/journal.pone.0045234
Chen L et al (2018) CD38-Mediated Immunosuppression as a Mechanism of Tumor Cell Escape from PD-1/PD-L1 Blockade. Cancer Discov 8(9):1156–1175. https://doi.org/10.1158/2159-8290.CD-17-1033
Rosser EC, Mauri C (2015) Regulatory B Cells: Origin, Phenotype, and Function. Immunity 42(4):607–612. https://doi.org/10.1016/j.immuni.2015.04.005
Chen PY, Wu CYJ, Fang JH, Chen HC, Feng LY, Huang CY, Wei KC, Fang JY, Lin CY (2019) Functional Change of Effector Tumor-Infiltrating CCR5(+)CD38(+)HLA-DR(+)CD8(+) T Cells in Glioma Microenvironment. Front Immunol 10:2395. https://doi.org/10.3389/fimmu.2019.02395
Flores-Borja F, Bosma A, Ng D, Reddy V, Ehrenstein MR, Isenberg DA, Mauri C (2013) CD19+CD24hiCD38hi B cells maintain regulatory T cells while limiting TH1 and TH17 differentiation. Sci Transl Med 5(173):173ra23. https://doi.org/10.1126/scitranslmed.3005407
Wang WW, Yuan XL, Chen H, Xie GH, Ma YH, Zheng YX, Zhou YL, Shen LS (2015) CD19+CD24hiCD38hiBregs involved in downregulate helper T cells and upregulate regulatory T cells in gastric cancer. Oncotarget 6(32):33486–33499. https://doi.org/10.18632/oncotarget.5588
Patton DT, Wilson MD, Rowan WC, Soond DR, Okkenhaug K (2011) The PI3K p110δ regulates expression of CD38 on regulatory T cells. PLoS One 6(3):e17359. https://doi.org/10.1371/journal.pone.0017359
Feng X, Zhang L, Acharya C, An G, Wen K, Qiu L, Munshi NC, Tai YT, Anderson KC (2017) Targeting CD38 Suppresses Induction and Function of T Regulatory Cells to Mitigate Immunosuppression in Multiple Myeloma. Clin Cancer Res 23(15):4290–4300. https://doi.org/10.1158/1078-0432.CCR-16-3192
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98–W102. https://doi.org/10.1093/nar/gkx247
Perenkov AD, Novikov DV, Sakharnov NA, Aliasova AV, Utkin OV, Baryshnikov AI, Novikov VV (2012) Heterogeneous expression of CD38 gene in tumor tissue in patients with colorectal cancer. Mol Biol (Mosk) 46(5):786–791
Chmielewski JP, Bowlby SC, Wheeler FB, Shi L, Sui G, Davis AL, Howard TD, D’Agostino RB, Miller LD, Sirintrapun SJ, Cramer SD, Kridel SJ (2020) CD38 Inhibits Prostate Cancer Metabolism and Proliferation by Reducing Cellular NAD(+) Pools. Mol Cancer Res 16(11):1687–1700. https://doi.org/10.1158/1541-7786.MCR-17-0526
Zhu Y, Zhang Z, Jiang Z, Liu, Y, Zhou J (2020) CD38 Predicts Favorable Prognosis by Enhancing Immune Infiltration and Antitumor Immunity in the Epithelial Ovarian Cancer Microenvironment. Front Genet 11. https://doi.org/10.3389/fgene.2020.00369
Rockenbach L, Braganhol E, Dietrich F, Figueiro F, Pugliesi M, Edelweiss MI, Morrone FB, Sévigny J, Battastini AMO (2014) NTPDase3 and ecto-5’-nucleotidase/CD73 are differentially expressed during mouse bladder cancer progression. Purinergic Signal 10(3):421–430. https://doi.org/10.1007/s11302-014-9405-8
Yu W, Robson SC, Hill WG (2011) Expression and distribution of ectonucleotidases in mouse urinary bladder. PLoS One 6(4):e18704. https://doi.org/10.1371/journal.pone.0018704
Huang J, Chen MN, Du J, Liu H, He YJ, Li GL, Li SY, Liu WP, Long XY (2016) Differential Expression of Adenosine P1 Receptor ADORA1 and ADORA2A Associated with Glioma Development and Tumor-Associated Epilepsy. Neurochem Res 41(7):1774–1783. https://doi.org/10.1007/s11064-016-1893-1
Bauer A, Langen KJ, Bidmon H, Holschbach MH, Weber S, Olsson RA, Coenen HH, Zilles K (2005) m18F-CPFPX PET identifies changes in cerebral A1 adenosine receptor density caused by glioma invasion. J Nucl Med 46(3):450–454
Takenaka MC, Gabriely G, Rothhammer V, Mascanfroni ID, Wheeler MA, Chao CC, Gutierrez-Vazquez C, Kenison J, Tjon EC, Barroso A, Vandeventer T, Lima KA, Rothweiler S, Mayo L, Ghannan S, Zandee S, Healy L, Sherr D, Farez MF, Prat A, Antel J, Reardon DA, Zhang H, Robson SC, Quintana FJ (2019) Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 22(5):729–740. https://doi.org/10.1038/s41593-019-0370-y
Yan A, Joachims ML, Thompson LF, Miller AD, Canoll PD, Bynoe MS (2019) CD73 Promotes Glioblastoma Pathogenesis and Enhances Its Chemoresistance via A(2B) Adenosine Receptor Signaling. J Neurosci Off J Soc Neurosci 39(22):4387–4402. https://doi.org/10.1523/JNEUROSCI.1118-18.2019
Bova V, Filippone A, Casili G, Lanza M, Compolo M, Capra AP, Repici A, Crupi L, Motta G, Colorossi C, Chisari G, Cuzzocrea S, Esposito E, Paterniti I (2022) Adenosine Targeting as a New Strategy to Decrease Glioblastoma Aggressiveness. Cancers (Basel) 14(16). https://doi.org/10.3390/cancers14164032
Kitabatake K, Kaji T, Tsukimoto M (2021) Involvement of CD73 and A2B Receptor in Radiation-Induced DNA Damage Response and Cell Migration in Human Glioblastoma A172 Cells. Biol Pharm Bull 44(2):197–210. https://doi.org/10.1248/bpb.b20-00654
Ott M, Tomaszowski KH, Marisetty A, Kong LY, Wei J, Duna M, Blumberg K, Ji X, Jacobs C, Fuller GN, Langford LA, Huse JT, Long JP, Hu J, Li S, Weinberg JS, Prabhu SS, Sawaya R, Ferguson S, Rao G, Lang FF, Curran MA, Heimberger AB (2020) Profiling of patients with glioma reveals the dominant immunosuppressive axis is refractory to immune function restoration. JCI insight 5(17). https://doi.org/10.1172/jci.insight.134386
Kohanbash G, Carrera DA, Shrivastav S, Ahn BJ, Jahan N, Mazor T, Chheda ZS, Downey KM, Watchmaker PB, Beppler C, Warta R, Amankulor NA, Herold-Mende C, Costello JF, Okada H (2017) Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest 127(4):425–1437. https://doi.org/10.1172/JCI90644
Notarangelo G et al (2022) Oncometabolite d-2HG alters T cell metabolism to impair CD8(+) T cell function. Science 377(6614):1519–1529. https://doi.org/10.1126/science.abj5104
Acknowledgements
The authors would like to thank all the patients who have agreed to participate in this study and their families. We also thank all from Departamento de Neurocirurgia team (Hospital Cristo Redentor) for their work and collaboration, and all our collaborators for the technical support provided. Finally, we thank the funding agencies Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and Instituto Nacional de Ciência e Tecnologia (INCT).
Funding
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; n° 406035/2021–0 and PQ n° 311580/2021–1, Figueiró F), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS/PQG grant no. 21/2551–0001972-7) and Instituto Nacional de Ciência e Tecnologia—INCT/CNPq/CAPES/FAPERGS grant no. 465671/2014–4.). This study was financed in part by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; 140977/2019–8).
Author information
Authors and Affiliations
Contributions
Conceptualization: Juliete Nathali Scholl, Fabrício Figueiró; Methodology: Juliete Nathali Scholl, Augusto Ferreira Weber, Camila Kehl Dias, Vinícius Pierdoná Lima, Lucas Kich Grun, Diego Zambonin, Eduardo Anzolin, Wanderson Willian Dos Santos Dias, Willian Pegoraro Kus; Validation: Juliete Nathali Scholl, Augusto Ferreira Weber, Camila Kehl Dias, Vinícius Pierdoná Lima; Formal analysis and Investigation: Juliete Nathali Scholl, Augusto Ferreira Weber, Camila Kehl Dias, Vinícius Pierdoná Lima Writing—original draft: Juliete Nathali Scholl; Writing—review & editing: Juliete Nathali Scholl, Augusto Ferreira Weber, Camila Kehl Dias, Vinícius Pierdoná Lima, Lucas Kich Grun, Diego Zambonin, Eduardo Anzolin, Wanderson Willian Dos Santos Dias, Willian Pegoraro Kus, Florencia Barbé-Tuana, Ana Maria Oliveira Battastini, Paulo Valdeci Worm, Fabrício Figueiró; Visualization: Juliete Nathali Scholl; Supervision: Fabrício Figueiró; Project administration: Juliete Nathali Scholl, Fabrício Figueiró; Funding acquisition: Juliete Nathali Scholl, Fabrício Figueiró.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Conflicts of interest
Juliete Nathali Scholl declares that she has no conflict of interest.
Augusto Ferreira Weber declares that he has no conflict of interest.
Camila Kehl Dias declares that she has no conflict of interest.
Vinícius Pierdoná Lima declares that he has no conflict of interest.
Lucas Kich Grun declares that he has no conflict of interest.
Diego Zambonin declares that he has no conflict of interest.
Eduardo Anzolin declares that he has no conflict of interest.
Wanderson Willian Dos Santos Dias declares that he has no conflict of interest.
Willian Pegoraro Kus declares that he has no conflict of interest.
Florencia Barbé-Tuana declares that she has no conflict of interest.
Ana Maria Oliveira Battastini declares that she has no conflict of interest.
Paulo Valdeci Worm declares that he has no conflict of interest.
Fabrício Figueiró declares that he has no conflict of interest.
Ethics approval
The study was conducted in accordance with the 1964 Helsinki Declaration, and the protocol was approved by the Ethics Committee of the Grupo Hospitalar Conceição and the Universidade Federal do Rio Grande do Sul (Project n°: 4.343.931, CAAE n°: 24997119.8.3002.5530; and Project number: 3.986.203, CAAE number: 24997119.8.0000.5347).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Supplementary Fig. 1
Plasmatic purine levels. Nucleotides (A - C) and nucleosides (D - G) levels in LGG, HGG IDH-Mut, and HGG IDH-WT patient plasma samples were evaluated by HPLC. Data represent a combination of experiments involving individual patients and are displayed as the mean ± SD (PNG 55 kb)

Supplementary Fig. 2
Kaplan-Meier analysis showing the influence of CD38 (A, D, G, J, M, and P), ENTPD1 (B, E, H, K, N, Q, S, and U), and NT5E (C, F, I, L, O, R, T, and V) gene expression of HGG and LGG in patients with high Treg, CD8+, CD19+, and NK cell gene signature (PNG 453 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Scholl, J.N., Weber, A.F., Dias, C.K. et al. Characterization of purinergic signaling in tumor-infiltrating lymphocytes from lower- and high-grade gliomas. Purinergic Signalling 20, 47–64 (2024). https://doi.org/10.1007/s11302-023-09931-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11302-023-09931-4