Abstract
Background
Single nucleotide polymorphisms influence the effects of tacrolimus and cyclosporine in renal transplants.
Aim
We set out to use machine learning algorithms (MLAs) to identify variables that predict the therapeutic effects and adverse events following tacrolimus and cyclosporine administration in renal transplant patients.
Method
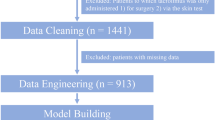
We sampled 120 adult renal transplant patients (on cyclosporine or tacrolimus). Generalized linear model (GLM), support vector machine (SVM), artificial neural network (ANN), Chi-square automatic interaction detection, classification and regression tree, and K-nearest neighbors were the chosen MLAs. The mean absolute error (MAE), relative mean square error (RMSE), and regression coefficient (β) with a 95% confidence interval (CI) were used as the model parameters.
Results
For a stable dose of tacrolimus, the MAEs (RMSEs) of GLM, SVM, and ANN were 1.3 (1.5), 1.3 (1.8), and 1.7 (2.3) mg/day, respectively. GLM revealed that the POR*28 genotype and age significantly predicted the stable dose of tacrolimus as follows: POR*28 (β −1.8; 95% CI −3, −0.5; p = 0.006), and age (β −0.04; 95% CI −0.1, −0.006; p = 0.02). For a stable dose of cyclosporine, MAEs (RMSEs) of 93.2 (103.4), 79.1 (115.2), and 73.7 (91.7) mg/day were observed with GLM, SVM, and ANN, respectively. GLM revealed the following predictors of a stable dose of cyclosporine: CYP3A5*3 (β −80.8; 95% CI −130.3, −31.2; p = 0.001), and age (β −3.4; 95% CI −5.9, −0.9; p = 0.007).
Conclusion
We observed that various MLAs could identify significant predictors that were useful to optimize tacrolimus and cyclosporine dosing regimens; yet, the findings must be externally validated.



Similar content being viewed by others
References
Meier-Kriesche HU, Li S, Gruessner RW, et al. Immunosuppression: evolution in practice and trends, 1994–2004. Am J Transplant. 2006;6(5 Pt 2):1111–31.
Thongprayoon C, Hansrivijit P, Kovvuru K, et al. Impacts of high intra- and inter-individual variability in tacrolimus pharmacokinetics and fast tacrolimus metabolism on outcomes of solid organ transplant recipients. J Clin Med. 2020;9(7):2193.
van Gelder T. Within-patient variability in immunosuppressive drug exposure as a predictor for poor outcome after transplantation. Kidney Int. 2014;85(6):1267–8.
Nobakht E, Jagadeesan M, Paul R, et al. Precision medicine in kidney transplantation: Just hype or a realistic hope? Transplant Direct. 2021;7(2): e650.
Lee DH, Lee H, Yoon HY, et al. Association of P450 oxidoreductase gene polymorphism with tacrolimus pharmacokinetics in renal transplant recipients: a systematic review and meta-analysis. Pharmaceutics. 2022;14(2):261.
Seyahi N, Ozcan SG. Artificial intelligence and kidney transplantation. World J Transplant. 2021;11(7):277–89.
Tang J, Liu R, Zhang YL, et al. Application of machine-learning models to predict tacrolimus stable dose in renal transplant recipients. Sci Rep. 2018;8:46936.
Gim JA, Kwon Y, Lee HA, et al. A machine learning-based identification of genes affecting the pharmacokinetics of tacrolimus using the DMETTM plus platform. Int J Mol Sci. 2020;21(7):2517.
Thishya K, Vattam KK, Naushad SM, et al. Artificial neural network model for predicting the bioavailability of tacrolimus in patients with renal transplantation. PLoS ONE. 2018;13(4):e0191921.
Zhou Y, Lauschke VM. Population pharmacogenomics: an update on ethnogeographic differences and opportunities for precision public health. Hum Genet. 2021;141:1113–36. https://doi.org/10.1007/s00439-021-02385-x.
Sridharan K, Shah S, Jassim A, et al. Evaluation of pharmacogenetics of drug-metabolizing enzymes and drug efflux transporter in renal transplants receiving immunosuppressants. J Pers Med. 2022;12(5):823.
Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. J Clin Epidemiol. 2015;68(2):134–43.
Mulder TAM, van Eerden RAG, de With M, et al. CYP3A4∗22 genotyping in clinical practice: Ready for implementation? Front Genet. 2021;12: 711943.
Saiz-Rodríguez M, Almenara S, Navares-Gómez M, et al. Effect of the most relevant CYP3A4 and CYP3A5 polymorphisms on the pharmacokinetic parameters of 10 CYP3A substrates. Biomedicines. 2020;8(4):94.
Li M, Xu M, Liu W, et al. Effect of CYP3 A4, CYP3 A5 and ABCB1 gene polymorphisms on the clinical efficacy of tacrolimus in the treatment of nephrotic syndrome. BMC Pharmacol Toxicol. 2018;19(1):14.
Chernov A, Kilina D, Smirnova T, et al. Pharmacogenetic study of the impact of ABCB1 single nucleotide polymorphisms on the response to cyclosporine in psoriasis patients. Pharmaceutics. 2022;14(11):2441.
Masters BS. The journey from NADPH-cytochrome P450 oxidoreductase to nitric oxide synthases. Biochem Biophys Res Commun. 2005;338(1):507–19.
Huang N, Agrawal V, Giacomini KM, et al. Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc Natl Acad Sci USA. 2008;105(5):1733–8.
Elens L, Nieuweboer AJ, Clarke SJ, et al. Impact of POR*28 on the clinical pharmacokinetics of CYP3A phenotyping probes midazolam and erythromycin. Pharmacogenet Genomics. 2013;23(3):148–55.
Tang JT, Andrews LM, van Gelder T, et al. Pharmacogenetic aspects of the use of tacrolimus in renal transplantation: recent developments and ethnic considerations. Expert Opin Drug Metab Toxicol. 2016;12(5):555–65.
Wang P, Mao Y, Razo J, et al. Using genetic and clinical factors to predict tacrolimus dose in renal transplant recipients. Pharmacogenomics. 2010;11(10):1389–402.
Wendler T, Grottrup S. Regression models. In: Data mining with SPSS modeler. Springer, Cham, Switzerland, pp 367–546.
Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287–98.
Zhang JJ, Liu SB, Xue L, et al. The genetic polymorphisms of POR*28 and CYP3A5*3 significantly influence the pharmacokinetics of tacrolimus in Chinese renal transplant recipients. Int J Clin Pharmacol Ther. 2015;53(9):728–36.
Chitnis SD, Ogasawara K, Schniedewind B, et al. Concentration of tacrolimus and major metabolites in kidney transplant recipients as a function of diabetes mellitus and cytochrome P450 3A gene polymorphism. Xenobiotica. 2013;43(7):641–9.
Funding
No specific funding was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sridharan, K., Shah, S. Developing supervised machine learning algorithms to evaluate the therapeutic effect and laboratory-related adverse events of cyclosporine and tacrolimus in renal transplants. Int J Clin Pharm 45, 659–668 (2023). https://doi.org/10.1007/s11096-023-01545-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11096-023-01545-5