Abstract
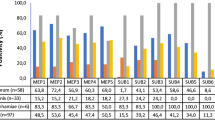
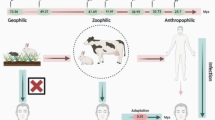
The two fungal species Trichophyton rubrum and Trichophyton violaceum are common pathogens on human, infecting keratinized tissue of the outer body parts. Both species are belonging to the “Trichophyton rubrum complex” and share very high similarity in the genome. Secreted proteinases, key factors for keratin degradation, are nearly identical. Contrary, the ecological niches are differing. Trichophyton rubrum preferably infects skin and nails, whereas T. violaceum preferably infects the scalp. We postulate, that differences in the protease expression contribute to differences in ecological preferences. We analyzed the expression profiles of all 22 endoprotease genes, 12 subtilisins (S8A), 5 deuterolysins (M35) and 5 fungalysins (M36), for both species. To compare the influence of the keratin source, we designed experiments with human nail keratin, sheep wool keratin and keratin free cultivation media. Samples were taken at 12 h, 24 h, 48 h and 96 h post incubation in keratin medium. The expression of the proteases is higher in wool-keratin medium compared to human nail medium, with the exception of MEP4 and SUB6. Expression in the keratin-free medium is lowest. The expression profiles of the two species are remarkable different. The expression of MEP1, MEP3, SUB5, SUB11 and SUB12 are higher in T. rubrum compared to T. violaceum. MEP2, NpIIc, NpIIe, SUB1, SUB3, SUB4, SUB7 and SUB8 are higher expressed in T. violaceum compared to T. rubrum. The differences of the protease expression in the two species may expalin the differences in the ecological niches. Further analysis are necessary to verify the hypothesis.Please check and conform the edit made in title.Here I thinke the species of strains shouldnt be capital, and the right expression should be, "Expression Profiles of Protease in Onychomycosis-Related Pathogenic Trichophyton rubrum and Tinea Capitis-Related Pathogenic Trichophyton violaceum"Author names: Please confirm if the author names are presented accurately and in the correct se-quence (given name, middle name/initial, family name). Author 1 Given name: [Jingjing] Last name [Chen], Author 2 Given name: [Yangmin] Last name [Gao], Author 3 Given name: [Shuzhen] Last name [Xiong], Author 4 Given name: [Ping] Last name [Zhan]. Also, kindly confirm the details in the metadata are correct.YesPlease check and confirm the inserted city and country are correctly identified for affiliation 3.Please change the affiliations, Affiliation 2: ²Jiangxi Provincial Clinical Research Center for Skin Diseases, Dermatology Hospital of Jiangxi Province,The Affiliated Dermatology Hospital of Nanchang University, Nanchang, 330200, Jiangxi; Affiliation 3: 3Institute of Clinical Medicine, Jiangxi Provincial People's Hospital, The First Affiliated Hospital of Nanchang Medical College,Nanchang 330001, Jiangxi. Thanks a lot!





Similar content being viewed by others
References
Lin BB, Pattle N, Kelley P, Jaksic AS. Multiplex RT-PCR provides improved diagnosis of skin and nail dermatophyte infections compared to microscopy and culture: a laboratory study and review of the literature. Diagn Microbiol Infect Dis. 2021;101(3): 115413. https://doi.org/10.1016/j.diagmicrobio.2021.115413.
Jousson O, Lechenne B, Bontems O, Capoccia S, Mignon B, Barblan J, et al. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology (Reading). 2004;150(Pt 2):301–10. https://doi.org/10.1099/mic.0.26690-0.
Elewski BE, Charif MA. Prevalence of onychomycosis in patients attending a dermatology clinic in northeastern Ohio for other conditions. Arch Dermatol. 1997;133(9):1172–3.
Nenoff P, Kruger C, Ginter-Hanselmayer G, Tietz HJ. Mycology–an update. Part 1: dermatomycoses: causative agents, epidemiology and pathogenesis. J Dtsch Dermatol Ges. 2014;12(3):188–209. https://doi.org/10.1111/ddg.12245.
Zaraa I, Hawilo A, Aounallah A, Trojjet S, El EuchMokni DM, et al. Inflammatory Tinea capitis: a 12-year study and a review of the literature. Mycoses. 2013;56(2):110–6. https://doi.org/10.1111/j.1439-0507.2012.02219.x.
Zhan P, Dukik K, Li D, Sun J, Stielow JB, Gerrits van den Ende B, et al. Phylogeny of dermatophytes with genomic character evaluation of clinically distinct Trichophyton rubrum and T violaceum. Stud Mycol. 2018;89:153–75. https://doi.org/10.1016/j.simyco.2018.02.004.
Mercer DK, Stewart CS. Keratin hydrolysis by dermatophytes. Med Mycol. 2019;57(1):13–22. https://doi.org/10.1093/mmy/myx160.
Samdani AJ, Dykes PJ, Marks R. The proteolytic activity of strains of T. mentagrophytes and T. rubrum isolated from tinea pedis and tinea unguium infections. J Med Vet Mycol. 1995;33(3):167–70.
Viani FC, Dos SantosPaula JICR, Larson CE, Gambale W. Production of extracellular enzymes by Microsporum canis and their role in its virulence. Med Mycol. 2001;39(5):463–8. https://doi.org/10.1080/mmy.39.5.463.468.
Monod M, Capoccia S, Lechenne B, Zaugg C, Holdom M, Jousson O. Secreted proteases from pathogenic fungi. Int J Med Microbiol. 2002;292(5–6):405–19. https://doi.org/10.1078/1438-4221-00223.
Monod M. Secreted proteases from dermatophytes. Mycopathologia. 2008;166(5–6):285–94. https://doi.org/10.1007/s11046-008-9105-4.
Li J, Zhang KQ. Independent expansion of zincin metalloproteinases in Onygenales fungi may be associated with their pathogenicity. PLoS One. 2014;9(2): e90225. https://doi.org/10.1371/journal.pone.0090225.
Yike I. Fungal proteases and their pathophysiological effects. Mycopathologia. 2011;171(5):299–323. https://doi.org/10.1007/s11046-010-9386-2.
Leng W, Liu T, Wang J, Li R, Jin Q. Expression dynamics of secreted protease genes in Trichophyton rubrum induced by key host’s proteinaceous components. Med Mycol. 2009;47(7):759–65. https://doi.org/10.3109/13693780802524522.
Apodaca G, McKerrow JH. Regulation of Trichophyton rubrum proteolytic activity. Infect Immun. 1989;57(10):3081–90. https://doi.org/10.1128/iai.57.10.3081-3090.1989.
Giddey K, Favre B, Quadroni M, Monod M. Closely related dermatophyte species produce different patterns of secreted proteins. FEMS Microbiol Lett. 2007;267(1):95–101. https://doi.org/10.1111/j.1574-6968.2006.00541.x.
Li J, Yu L, Tian Y, Zhang KQ. Molecular evolution of the deuterolysin (M35) family genes in Coccidioides. PLoS One. 2012;7(2): e31536. https://doi.org/10.1371/journal.pone.0031536.
Chen J, Yi J, Liu L, Yin S, Chen R, Li M, et al. Substrate adaptation of Trichophyton rubrum secreted endoproteases. Microb Pathog. 2010;48(2):57–61. https://doi.org/10.1016/j.micpath.2009.12.001.
Bruce Fraser RD, Parry DA. The role of disulfide bond formation in the structural transition observed in the intermediate filaments of developing hair. J Struct Biol. 2012;180(1):117–24. https://doi.org/10.1016/j.jsb.2012.05.020.
Gong H, Zhou H, McKenzie GW, Yu Z, Clerens S, Dyer JM, et al. An updated nomenclature for keratin-associated proteins (KAPs). Int J Biol Sci. 2012;8(2):258–64. https://doi.org/10.7150/ijbs.3278.
Bitencourt TA, Macedo C, Franco ME, Assis AF, Komoto TT, Stehling EG, et al. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. BMC Genomics. 2016;17:249. https://doi.org/10.1186/s12864-016-2567-8.
Tran VD, De CoiFeuermann NM, Schmid-Siegert E, Bagut ET, Mignon B, et al. RNA sequencing-based genome reannotation of the dermatophyte Arthroderma benhamiae and characterization of its secretome and whole gene expression profile during infection. mSystems. 2016. https://doi.org/10.1128/mSystems.00036-16.
Acknowledgements
No further acknowledgments.
Funding
This work was supported by the Key Scientific Project of Administration of Traditional Chinese Medicine of Jiangxi Province (2023Z008) and the National Natural Science Foundation of China [81960367], the Key Research and Development Project of Jiangxi Province (20203BBG73040), National Natural Science Foundation of Jiangxi Province [20212BAB206060] and Science and Technology innovation Team of Jiangxi University of Chinese Medicine (#CXTD22009).
Author information
Authors and Affiliations
Contributions
CJ and GY perform the experiment and original writing, XS and PZ help with data analysis, and PZ proposed the study and contributed to the writing polishing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Handling Editor: Ruoyu Li.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J., Gao, Y., Xiong, S. et al. Expression Profiles of Protease in Onychomycosis-Related Pathogenic Trichophyton rubrum and Tinea Capitis-Related Pathogenic Trichophyton violaceum. Mycopathologia 189, 14 (2024). https://doi.org/10.1007/s11046-024-00828-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11046-024-00828-3