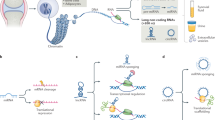
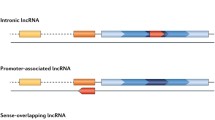
Abstract
Inflammation and autoimmunity are the root cause of rheumatoid arthritis, a destructive disease of joints. Multiple biomolecules are involved in the pathogenesis of RA and are related to various events of molecular biology. RNA is a versatile biomolecule, playing numerous roles at structural, functional, and regulatory stages to maintain cellular homeostasis. The involvement of RNA (coding/non-coding) in disease development and progression has left a wide whole to fill with newer approaches. Non-coding RNAs belong to the housekeeping and regulatory categories and both have their specific roles, and their alteration causes specific implications in disease pathogenesis. Housekeeping RNAs, rRNA, tRNA and regulatory RNA, micro-RNA, circular RNA, piRNA and long non-coding RNA were found to be important regulators of inflammation. They work at the pre-and post-transcriptional levels and were found to be more intriguing to study their regulatory impact on disease pathogenesis. The review addresses a question on how the non-coding RNA gets involved in early RA pathogenesis and can be utilized to know their targets to understand the disease better and make way towards the unresolved mystery of RA development.

Similar content being viewed by others
Data Availability
Not applicable.
References
Siomi H, Siomi MC (2009) On the road to reading the RNA-interference code. Nature 457:396–404. https://doi.org/10.1038/NATURE07754
Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10:94–108. https://doi.org/10.1038/NRG2504
Statello L, Guo CJ, Chen LL, Huarte M (2020) Gene regulation by long non-coding RNAs and its biological functions. Nature Reviews Molecular Cell Biology 2020 22:2 22:96–118. https://doi.org/10.1038/s41580-020-00315-9
Brosnan CA, Voinnet O (2009) The long and the short of noncoding RNAs. Curr Opin Cell Biol 21:416–425. https://doi.org/10.1016/J.CEB.2009.04.001
Guo Q, Wang Y, Xu D et al (2018) Rheumatoid arthritis: pathological mechanisms and modern pharmacologic therapies. Bone Res 6. https://doi.org/10.1038/S41413-018-0016-9
Atzeni F, Talotta R, Masala IF et al (2021) Biomarkers in rheumatoid arthritis. Cureus 13:512–516. https://doi.org/10.7759/CUREUS.15063
Benjamin O, Bansal P, Goyal A, Lappin SL (2021) Disease Modifying Anti-Rheumatic Drugs (DMARD). StatPearls
Lee TI, Young RA (2013) Transcriptional regulation and its Misregulation in Disease. Cell 152:1237. https://doi.org/10.1016/J.CELL.2013.02.014
Chen W, Liu D, Li QZ, Zhu H (2019) The function of ncRNAs in rheumatic diseases. Epigenomics 11:821. https://doi.org/10.2217/EPI-2018-0135
Wang J, Yan S, Yang J et al (2019) Non-coding RNAs in rheumatoid arthritis: from bench to Bedside. Front Immunol 10:3129. https://doi.org/10.3389/FIMMU.2019.03129
Saikia M, Jobava R, Parisien M et al (2014) Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 34:2450. https://doi.org/10.1128/MCB.00136-14
Anderson P, Ivanov P (2014) tRNA fragments in human health and disease. FEBS Lett 588:4297. https://doi.org/10.1016/J.FEBSLET.2014.09.001
Yamasaki S, Nakashima M, Ida H (2021) Possible roles of tRNA fragments, as New Regulatory ncRNAs, in the pathogenesis of rheumatoid arthritis. Int J Mol Sci 2021 22:9481. https://doi.org/10.3390/IJMS22179481
Sarangdhar MA, Allam R (2021) Angiogenin (ANG)—Ribonuclease inhibitor (RNH1) system in protein synthesis and disease. Int J Mol Sci 22:1–11. https://doi.org/10.3390/IJMS22031287
Szekanecz Z, Besenyei T, Paragh G, Koch AE (2009) Angiogenesis in rheumatoid arthritis. Autoimmunity 42:563. https://doi.org/10.1080/08916930903143083
Magee R, Rigoutsos I (2020) On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res 48:9433–9448. https://doi.org/10.1093/NAR/GKAA657
Naddeo M, Vitagliano L, Russo A et al (2005) Interactions of the cytotoxic RNase a dimers with the cytosolic ribonuclease inhibitor. FEBS Lett 579:2663–2668. https://doi.org/10.1016/J.FEBSLET.2005.03.087
Yamasaki S, Ivanov P, Hu GF, Anderson P (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185:35–42. https://doi.org/10.1083/JCB.200811106
Kharel P, Becker G, Tsvetkov V, Ivanov P (2020) Properties and biological impact of RNA G-quadruplexes: from order to turmoil and back. Nucleic Acids Res 48:12534–12555. https://doi.org/10.1093/NAR/GKAA1126
Alberts B, Johnson A, Lewis J et al (2002) From RNA to Protein
Henras AK, Plisson-Chastang C, O’Donohue MF et al (2015) An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6:225. https://doi.org/10.1002/WRNA.1269
Luan M, Shang Z, Teng Y et al (2017) The shared and specific mechanism of four autoimmune diseases. Oncotarget 8:108355. https://doi.org/10.18632/ONCOTARGET.19383
Zhang P, Wu W, Chen Q, Chen M (2019) Non-coding RNAs and their Integrated Networks. J Integr Bioinform 16. https://doi.org/10.1515/JIB-2019-0027
Pratt AJ, MacRae IJ (2009) The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem 284:17897–17901. https://doi.org/10.1074/JBC.R900012200
Peters L, Meister G (2007) Argonaute proteins: mediators of RNA silencing. Mol Cell 26:611–623. https://doi.org/10.1016/J.MOLCEL.2007.05.001
Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10:126–139. https://doi.org/10.1038/NRM2632
Sala-Cirtog M, Marian C, Anghel A (2015) New insights of medicinal plant therapeutic activity-the miRNA transfer. Biomed Pharmacother 74:228–232. https://doi.org/10.1016/J.BIOPHA.2015.08.016
O’Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9:402. https://doi.org/10.3389/FENDO.2018.00402/BIBTEX
Condrat CE, Thompson DC, Barbu MG et al (2020) miRNAs as biomarkers in Disease: latest findings regarding their role in diagnosis and prognosis. Cells 9. https://doi.org/10.3390/CELLS9020276
Grimson A, Farh KKH, Johnston WK et al (2007) MicroRNA Targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27:91. https://doi.org/10.1016/J.MOLCEL.2007.06.017
Cortez MA, Bueso-Ramos C, Ferdin J et al (2011) MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol 8:467. https://doi.org/10.1038/NRCLINONC.2011.76
Riffo-Campos ÁL, Riquelme I, Brebi-Mieville P (2016) Tools for sequence-based miRNA target prediction: what to choose? Int J Mol Sci 17. https://doi.org/10.3390/IJMS17121987
Wang X, Xu X, Ma Z et al (2011) Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA 17:1511. https://doi.org/10.1261/RNA.2732611
Castro-Villegas C, Pérez-Sánchez C, Escudero A et al (2015) Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFα. Arthritis Res Ther 17. https://doi.org/10.1186/S13075-015-0555-Z
Ceribelli A, Nahid MA, Satoh M, Chan EKL (2011) MicroRNAs in rheumatoid arthritis. FEBS Lett 585:3667. https://doi.org/10.1016/J.FEBSLET.2011.05.020
Filková M, Aradi B, Šenolt L et al (2014) Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann Rheum Dis 73:1898–1904. https://doi.org/10.1136/ANNRHEUMDIS-2012-202815
Philippe L, Alsaleh G, Bahram S et al (2013) The miR-17 ∼ 92 cluster: a key player in the control of inflammation during rheumatoid arthritis. Front Immunol 4. https://doi.org/10.3389/FIMMU.2013.00070
Philippe L, Alsaleh G, Bahram S et al (2013) The miR-17 ~ 92 cluster: a key player in the control of inflammation during rheumatoid arthritis. Front Immunol 4. https://doi.org/10.3389/FIMMU.2013.00070/ABSTRACT
Peng WX, Zhu SL, Zhang BY et al (2017) Smoothened regulates Migration of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via activation of rho GTPase signaling. Front Immunol 8:159. https://doi.org/10.3389/FIMMU.2017.00159
Vega-Tapia F, Bustamante M, Valenzuela RA et al (2021) miRNA Landscape in Pathogenesis and Treatment of Vogt–Koyanagi–Harada Disease. Front Cell Dev Biol 9:863. https://doi.org/10.3389/FCELL.2021.658514/BIBTEX
Hong W, Zhang P, Wang X et al (2018) The Effects of MicroRNAs on Key Signalling Pathways and Epigenetic Modification in Fibroblast-Like Synoviocytes of Rheumatoid Arthritis. https://doi.org/10.1155/2018/9013124. Mediators Inflamm 2018:
Xiong G, Huang Z, Jiang H et al (2016) Inhibition of microRNA-21 decreases the invasiveness of fibroblast-like synoviocytes in rheumatoid arthritis via TGFβ/Smads signaling pathway. Iran J Basic Med Sci 19:787. https://doi.org/10.22038/ijbms.2016.7365
Li HW, Zeng HS (2020) Regulation of JAK/STAT signal pathway by miR-21 in the pathogenesis of juvenile idiopathic arthritis. World J Pediatr 16:502. https://doi.org/10.1007/S12519-019-00268-W
Lin J, Huo R, Xiao L et al (2014) A novel p53/microRNA-22/Cyr61 axis in synovial cells regulates inflammation in rheumatoid arthritis. Arthritis Rheumatol 66:49–59. https://doi.org/10.1002/ART.38142
Liu X, Ni S, Li C et al (2019) Circulating MicroRNA-23b as a New Biomarker for Rheumatoid Arthritis. Gene 712:143911. https://doi.org/10.1016/J.GENE.2019.06.001
Bordon Y (2012) MicroRNA-23b keeps TABs on tissue inflammation. Nature Reviews Immunology 2012 12:7 12:475–475. https://doi.org/10.1038/nri3250
Hou C, Wang D, Zhang L (2019) MicroRNA-34a-3p inhibits proliferation of rheumatoid arthritis fibroblast-like synoviocytes. Mol Med Rep 20:2563. https://doi.org/10.3892/MMR.2019.10516
Wu H, Zhou X, Wang X et al (2021) miR-34a in extracellular vesicles from bone marrow mesenchymal stem cells reduces rheumatoid arthritis inflammation via the cyclin I/ATM/ATR/p53 axis. J Cell Mol Med 25:1896–1910. https://doi.org/10.1111/JCMM.15857
Di Martino MT, Leone E, Amodio N et al (2012) Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res 18:6260–6270. https://doi.org/10.1158/1078-0432.CCR-12-1708
Sanchita, Trivedi R, Asif MH, Trivedi PK (2018) Dietary plant miRNAs as an augmented therapy: cross-kingdom gene regulation. RNA Biol 15:1433. https://doi.org/10.1080/15476286.2018.1551693
Saquib M, Agnihotri P, Monu, Biswas S (2021) Exogenous miRNA: a perspective role as therapeutic in rheumatoid arthritis. Curr Rheumatol Rep 23. https://doi.org/10.1007/S11926-021-01009-7
Huang RY, Wu JQ, Liu ZH, Sun SL (2019) MicroRNAs in rheumatoid arthritis: what is the latest with regards to diagnostics? https://doi.org/101080/1473715920191599716 19. 363–366. https://doi.org/10.1080/14737159.2019.1599716
Murata K, Furu M, Yoshitomi H et al (2013) Comprehensive microRNA Analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS ONE 8. https://doi.org/10.1371/JOURNAL.PONE.0069118
Lu LF, Boldin MP, Chaudhry A et al (2010) Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142:914–929. https://doi.org/10.1016/J.CELL.2010.08.012
Salzman J (2016) Circular RNA expression: its potential regulation and function. Trends Genet 32:309–316. https://doi.org/10.1016/J.TIG.2016.03.002
Petkovic S, Müller S (2015) RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 43:2454–2465. https://doi.org/10.1093/NAR/GKV045
Yu J, Xie D, Huang N, Zhou Q (2021) Circular RNAs as novel diagnostic biomarkers and therapeutic targets in kidney disease. Front Med (Lausanne) 8:1536. https://doi.org/10.3389/FMED.2021.714958/BIBTEX
Jeck WR, Sorrentino JA, Wang K et al (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. https://doi.org/10.1261/RNA.035667.112
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:1–14. https://doi.org/10.1186/S13059-014-0409-Z/FIGURES/6
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. https://doi.org/10.1038/NATURE11993
Chen CY, Sarnow P (1995) Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–417. https://doi.org/10.1126/SCIENCE.7536344
Westholm JO, Miura P, Olson S et al (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9:1966–1980. https://doi.org/10.1016/J.CELREP.2014.10.062
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:1–14. https://doi.org/10.1186/S13059-014-0409-Z/FIGURES/6
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340. https://doi.org/10.1038/280339A0
Salzman J, Gawad C, Wang PL et al (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7:e30733. https://doi.org/10.1371/JOURNAL.PONE.0030733
Rybak-Wolf A, Stottmeister C, Glažar P et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. https://doi.org/10.1016/J.MOLCEL.2015.03.027
Wang F, Nazarali AJ, Ji S (2016) Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res 6:1167
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20:1829–1842. https://doi.org/10.1261/RNA.047126.114
Yu CY, Kuo HC (2019) The emerging roles and functions of circular RNAs and their generation. Journal of Biomedical Science 2019 26:1 26:1–12. https://doi.org/10.1186/S12929-019-0523-Z
Geng X, Jia Y, Zhang Y et al (2020) Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 12:267–283. https://doi.org/10.2217/EPI-2019-0295. /ASSET/IMAGES/LARGE/Fig. 2.JPEG
Xiang D, Li Y, Lin Y (2020) Circular RNA circCCDC66 contributes to malignant phenotype of Osteosarcoma by sponging mir-338-3p to Upregulate the expression of PTP1B. https://doi.org/10.1155/2020/4637109. Biomed Res Int 2020:
Holdt LM, Stahringer A, Sass K et al (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016 7(1 7):1–14. https://doi.org/10.1038/ncomms12429
Wang YY, Wang YZ, Zhang HY, He ZY (2021) The role of circular RNAs in brain and stroke. Front Bioscience - Landmark 26:36–50. https://doi.org/10.52586/4923
Jahani S, Nazeri E, Majidzadeh-A K et al (2020) Circular RNA; a new biomarker for breast cancer: a systematic review. J Cell Physiol 235:5501–5510. https://doi.org/10.1002/JCP.29558
Cheng D, Wang J, Dong Z, Li X (2021) Cancer-related circular RNA: diverse biological functions. Cancer Cell International 2021 21:1 21:1–16. https://doi.org/10.1186/S12935-020-01703-Z
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388. https://doi.org/10.1080/15476286.2015.1020271
Zhang X, Yang D, Wei Y (2018) Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther 11:3979–3987. https://doi.org/10.2147/OTT.S158316
Zhang R, Xu J, Zhao J, Wang X (2018) Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci 22:118–126. https://doi.org/10.26355/EURREV_201801_14108
Zhong Z, Huang M, Lv M et al (2017) Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 403:305–317. https://doi.org/10.1016/J.CANLET.2017.06.027
Meng J, Chen S, Han JX et al (2018) Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in Hepatocellular Carcinoma. Cancer Res 78:4150–4162. https://doi.org/10.1158/0008-5472.CAN-17-3009
Xu Y, Chen F (2021) Current status of functional studies on circular RNAs in rheumatoid arthritis and their potential role as diagnostic biomarkers. 14. https://doi.org/10.2147/JIR.S302846
Yang J, Cheng M, Gu B et al (2020) CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis 11. https://doi.org/10.1038/S41419-020-03038-Z
Zhou JD, Li XX, Zhang TJ et al (2019) circFADS2 protects LPS-treated chondrocytes from apoptosis acting as an interceptor of miR-498/mTOR cross-talking. Aging 11:3348. https://doi.org/10.18632/AGING.101986
Chen G, Tang W, Wang S et al (2021) Promising diagnostic and therapeutic circRNAs for skeletal and chondral disorders. Int J Biol Sci 17:1428. https://doi.org/10.7150/IJBS.57887
Zhong S, Ouyang Q, Zhu D et al (2020) Hsa_circ_0088036 promotes the proliferation and migration of fibroblast-like synoviocytes by sponging mir-140-3p and upregulating SIRT 1 expression in rheumatoid arthritis. Mol Immunol 125:131–139. https://doi.org/10.1016/J.MOLIMM.2020.07.004
Zheng F, Yu X, Huang J, Dai Y (2017) Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep 16:8029–8036. https://doi.org/10.3892/MMR.2017.7638
Yang Q, Li R, Lyu Q et al (2019) Single-cell CAS-seq reveals a class of short PIWI-interacting RNAs in human oocytes. Nat Commun 2019 10:1. https://doi.org/10.1038/s41467-019-11312-8
Wu L, Jiang Y, Zheng Z et al (2020) mRNA and P-element-induced wimpy testis-interacting RNA profile in chemical-induced oral squamous cell carcinoma mice model. Exp Anim 69:168–177. https://doi.org/10.1538/EXPANIM.19-0042
Meseure D, Vacher S, Boudjemaa S et al (2020) Biopathological significance of PIWI–piRNA pathway deregulation in invasive breast carcinomas. Cancers (Basel) 12:1–21. https://doi.org/10.3390/CANCERS12102833
Tóth KF, Pezic D, Stuwe E, Webster A (2016) The piRNA Pathway Guards the germline genome against transposable elements. Adv Exp Med Biol 886:51. https://doi.org/10.1007/978-94-017-7417-8_4
Wu X, Pan Y, Fang Y et al (2020) The Biogenesis and Functions of piRNAs in Human Diseases. 21. https://doi.org/10.1016/J.OMTN.2020.05.023
Huang X, Fejes Tóth K, Aravin AA (2017) piRNA Biogenesis in Drosophila Melanogaster. Trends Genet 33:882. https://doi.org/10.1016/J.TIG.2017.09.002
Weick EM, Miska EA (2014) piRNAs: from biogenesis to function. Development 141:3458–3471. https://doi.org/10.1242/DEV.094037
Bochyńska A, Lüscher-Firzlaff J, Lüscher B (2018) Modes of Interaction of KMT2 histone H3 lysine 4 Methyltransferase/COMPASS complexes with chromatin. https://doi.org/10.3390/CELLS7030017. Cells 7:
Chen YCA, Stuwe E, Luo Y et al (2016) Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol Cell 63:97–109. https://doi.org/10.1016/J.MOLCEL.2016.05.010
Ge DT, Wang W, Tipping C et al (2019) The RNA-Binding ATPase, Armitage, couples piRNA amplification in nuage to phased piRNA production on Mitochondria. Mol Cell 74:982–995e6. https://doi.org/10.1016/J.MOLCEL.2019.04.006
Abe M, Naqvi A, Hendriks GJ et al (2014) Impact of age-associated increase in 2′-O-methylation of miRNAs on aging and neurodegeneration in Drosophila. Genes Dev 28:44. https://doi.org/10.1101/GAD.226654.113
Wang W, Han BW, Tipping C et al (2015) Slicing and binding by Ago3 or aub trigger piwi-bound piRNA production by distinct mechanisms. Mol Cell 59:819. https://doi.org/10.1016/J.MOLCEL.2015.08.007
Malone CD, Hannon GJ (2009) Molecular evolution of piRNA and Transposon Control Pathways in Drosophila. Cold Spring Harb Symp Quant Biol 74:225. https://doi.org/10.1101/SQB.2009.74.052
Watanabe T, Lin H (2014) Post-transcriptional regulation of gene expression by Piwi proteins and piRNAs. Mol Cell 56:18. https://doi.org/10.1016/J.MOLCEL.2014.09.012
Yu Y, Xiao J, Hann SS (2019) The emerging roles of PIWI-interacting RNA in human cancers. Cancer Manag Res 11:5895. https://doi.org/10.2147/CMAR.S209300
Han Y, Neng;, Li Y et al (2017) PIWI proteins and PIWI-Interacting RNA: emerging roles in Cancer. Cell Physiol Biochem 44:1–20. https://doi.org/10.1159/000484541
Pleštilová L, Neidhart M, Russo G et al (2016) Expression and regulation of PIWIL-Proteins and PIWI-Interacting RNAs in rheumatoid arthritis. PLoS ONE 11. https://doi.org/10.1371/JOURNAL.PONE.0166920
Sakuraba K, Fujimura K, Kamura S, IN B CELLS IS CRITICAL FOR THE DEVELOPMENT OF COLLAGEN INDUCED ARTHRITIS IN MICE THU0056 EXPRESSION OF PIWI-INTERACTING RNA IN RHEUMATOID (2007) THU0055 IL-21 SIGNALING. J Exp Med 448:365–378. https://doi.org/10.1136/annrheumdis-2015-eular.3434
Gesualdo F, Di, Capaccioli S, Lulli M (2014) A pathophysiological view of the long non-coding RNA world. Oncotarget 5:10976. https://doi.org/10.18632/ONCOTARGET.2770
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10:924. https://doi.org/10.4161/RNA.24604
Derrien T, Johnson R, Bussotti G et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775. https://doi.org/10.1101/GR.132159.111
Kung JTY, Colognori D, Lee JT (2013) Long noncoding RNAs: past, Present, and Future. Genetics 193:651. https://doi.org/10.1534/GENETICS.112.146704
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904. https://doi.org/10.1016/J.MOLCEL.2011.08.018
Bhat SA, Ahmad SM, Mumtaz PT et al (2016) Long non-coding RNAs: mechanism of action and functional utility. Noncoding RNA Res 1:43. https://doi.org/10.1016/J.NCRNA.2016.11.002
Ahmad P, Bensaoud C, Mekki I et al (2021) Long Non-Coding RNAs and Their Potential Roles in the Vector–Host–Pathogen Triad. Life 2021, Vol 11, Page 56 11:56. https://doi.org/10.3390/LIFE11010056
Sánchez Y, Huarte M (2013) Long non-coding RNAs: Challenges for diagnosis and therapies. Nucleic Acid Ther 23:15. https://doi.org/10.1089/NAT.2012.0414
Miao C, Bai L, Yang Y, Huang J (2021) Dysregulation of lncRNAs in rheumatoid arthritis: biomarkers, pathogenesis and potential therapeutic targets. Front Pharmacol 12. https://doi.org/10.3389/FPHAR.2021.652751
Mo BY, Guo XH, Yang MR et al (2018) Long non-coding RNA GAPLINC promotes Tumor-Like Biologic Behaviors of Fibroblast-Like Synoviocytes as MicroRNA Sponging in Rheumatoid Arthritis Patients. Front Immunol 9. https://doi.org/10.3389/FIMMU.2018.00702
Cantile M, di Bonito M, de Bellis MT, Botti G (2021) Functional Interaction among lncRNA HOTAIR and MicroRNAs in Cancer and Other Human Diseases. Cancers (Basel) 13:1–22. https://doi.org/10.3390/CANCERS13030570
Yuan M, Wang S, Yu L et al (2017) Long noncoding RNA profiling revealed differentially expressed lncRNAs associated with disease activity in PBMCs from patients with rheumatoid arthritis. PLoS ONE 12. https://doi.org/10.1371/JOURNAL.PONE.0186795
Liang J, Chen W, Lin DrJ (2019) LncRNA: an All-rounder in Rheumatoid Arthritis. J Transl Int Med 7:3. https://doi.org/10.2478/JTIM-2019-0002
Jash A, Sahoo A, Kim GC et al (2012) Nuclear factor of activated T cells 1 (NFAT1)-induced permissive chromatin modification facilitates nuclear factor-κB (NF-κB)-mediated interleukin-9 (IL-9) transactivation. J Biol Chem 287:15445–15457. https://doi.org/10.1074/JBC.M112.340356
Amirinejad R, Rezaei M, Shirvani-Farsani Z (2020) An update on long intergenic noncoding RNA p21: a regulatory molecule with various significant functions in cancer. Cell Biosci 10. https://doi.org/10.1186/S13578-020-00445-9
Liu Y, Yang L, Xu Q et al (2019) Long noncoding RNA MEG3 regulates rheumatoid arthritis by targeting NLRC5. J Cell Physiol 234:14270–14284. https://doi.org/10.1002/JCP.28126
Zou Y, Xu S, Xiao Y et al (2018) Long noncoding RNA LERFS negatively regulates rheumatoid synovial aggression and proliferation. J Clin Invest 128:4510. https://doi.org/10.1172/JCI97965
Geng X, Xu X, Fang Y et al (2019) Effect of long non-coding RNA growth arrest-specific 5 on apoptosis in renal ischaemia/reperfusion injury. Nephrol (Carlton) 24:405–413. https://doi.org/10.1111/NEP.13476
Song J, Kim D, Han J et al (2015) PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med 15:121–126. https://doi.org/10.1007/S10238-013-0271-4
Ye Y, Gao X, Yang N (2018) LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Hum Cell 31:14–21. https://doi.org/10.1007/S13577-017-0179-5
Natarelli L, Weber C (2022) A non-canonical link between non-coding RNAs and Cardiovascular Diseases. https://doi.org/10.3390/BIOMEDICINES10020445. Biomedicines 10:
Cole C, Sobala A, Lu C et al (2009) Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small RNAs derived from tRNAs. RNA 15:2147. https://doi.org/10.1261/RNA.1738409
Ormseth MJ, Solus JF, Sheng Q et al (2020) The endogenous plasma small RNAome of rheumatoid arthritis. ACR Open Rheumatol 2:97. https://doi.org/10.1002/ACR2.11098
Guglas K, Kołodziejczak I, Kolenda T et al (2020) YRNAs and YRNA-Derived fragments as New Players in Cancer Research and their potential role in Diagnostics. Int J Mol Sci 21:1–12. https://doi.org/10.3390/IJMS21165682
Stavast CJ, Erkeland SJ (2019) The non-canonical aspects of MicroRNAs: many roads to Gene Regulation. https://doi.org/10.3390/CELLS8111465. Cells 8:
Dika E, Broseghini E, Porcellini E et al (2021) Unraveling the role of microRNA/isomiR network in multiple primary melanoma pathogenesis. Cell Death Dis 12. https://doi.org/10.1038/S41419-021-03764-Y
Rorbach G, Unold O, Konopka BM (2018) Distinguishing mirtrons from canonical miRNAs with data exploration and machine learning methods. Sci Rep 8. https://doi.org/10.1038/S41598-018-25578-3
Naganuma T, Hirose T (2013) Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol 10:456. https://doi.org/10.4161/RNA.23547
Geng X, Jia Y, Zhang Y et al (2020) Circular RNA: biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 12:267–283. https://doi.org/10.2217/EPI-2019-0295
López-Jiménez E, Andrés-León E (2021) The Implications of ncRNAs in the Development of Human Diseases. Non-Coding RNA 2021, Vol 7, Page 17 7:17. https://doi.org/10.3390/NCRNA7010017
Afrashteh Nour M, Ghorbaninezhad F, Asadzadeh Z et al (2023) The emerging role of noncoding RNAs in systemic lupus erythematosus: new insights into the master regulators of disease pathogenesis. Ther Adv Chronic Dis 14. https://doi.org/10.1177/20406223231153572
Heydari R, Koohi F, Rasouli M et al (2023) Exosomes as Rheumatoid Arthritis Diagnostic Biomarkers and Therapeutic Agents. Vaccines (Basel) 11:. https://doi.org/10.3390/VACCINES11030687
Lu H, Yang Y, Kuang D et al (2022) Expression profile of circRNA in peripheral blood mononuclear cells of patients with rheumatoid arthritis. BMC Med Genomics 15. https://doi.org/10.1186/S12920-022-01225-9
Jia DD, Jiang H, Zhang YF et al (2022) The regulatory function of piRNA/PIWI complex in cancer and other human diseases: the role of DNA methylation. Int J Biol Sci 18:3358. https://doi.org/10.7150/IJBS.68221
Acknowledgements
We acknowledge Council of Scientific and Industrial Research (CSIR), Department of Science and Technology (DST), Government of India, New Delhi, India for providing financial support. Mohd Saquib received fellowship support from CSIR, Prachi Agnihotri received a fellowship from DST project GAP0212, and Monu received fellowship support from CSIR. We also thank Council of Scientific and Industrial Research (CSIR)-Institute of Genomics and Integrative Biology, Delhi, India for research and AcSIR for academic support.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to data analysis and took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal. Mohd Saquib and Dr Sagarika Biswas made substantial contributions to conception, design, drafting, acquisition of data, or analysis.
Corresponding author
Ethics declarations
Conflict of interest / Competing interests
All authors declared no conflict of interest.
Ethical approval
and Consent to participate:
Not applicable.
Human and Animal Ethics
Not applicable.
Consent for publication
All of the authors have agreed that the study should be submitted to Functional & Integrative Genomics.
Authors’ information
Council of Scientific and Industrial Research (CSIR)-Institute of Genomics and Integrative Biology, Mall Road, Delhi University Campus, Delhi, India,110007.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saquib, M., Agnihotri, P., Monu et al. Interrelated grid of non-coding RNA: An important aspect in Rheumatoid Arthritis pathogenesis. Mol Biol Rep 50, 6217–6232 (2023). https://doi.org/10.1007/s11033-023-08543-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08543-w