Abstract
Background
Vegetable soybean seeds are among the most popular and nutrient-dense beans in the world due to their delicious flavor, high yield, superior nutritional value, and low trypsin content. There is significant potential for this crop that Indian farmers do not fully appreciate because of the limited germplasm range. Therefore, the current study aims to identify the diverse lines of vegetable soybean and explore the diversity produced by hybridizing grain and vegetable-type soybean varieties. Indian researchers have not yet published work describing and analysing novel vegetable soybean for microsatellite markers and morphological traits.
Methods and results
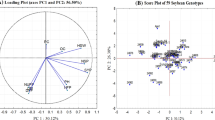
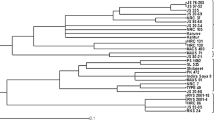
Sixty polymorphic SSR markers and 19 morphological traits were used to evaluate the genetic diversity of 21 newly developed vegetable soybean genotypes. A total of 238 alleles, ranging from 2 to 8, were found, with a mean of 3.97 alleles per locus. The polymorphism information content varied from 0.05 to 0.85, with an average of 0.60. A variation of 0.25–0.58 with a mean of 0.43 was observed for Jaccard’s dissimilarity coefficient.
Conclusion
The diverse genotypes identified can be helpful to understand the genetics of vegetable soybean traits and can be used in improvement programs; study also explains the utility of SSR markers for diversity analysis of vegetable soybean. Here, we identified the highly informative SSRs with PIC > 0.80 (satt199, satt165, satt167, satt191, satt183, satt202, and satt126), which apply to genetic structure analysis, mapping strategies, polymorphic marker surveys, and background selection in genomics-assisted breeding.






Similar content being viewed by others
Data availability
The data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Esler I (2011) Prospects for Vegetable Soybean in India and its Market Acceptance. Research and Cultural Experiences in Hyderabad, India. https://www.worldfoodprize.org/documents/filelibrary/images/youth_programs/2011_interns/EslerIzzy_India_DD3E1352FCBE2.pdf Accessed on Dec 2021.
Mathur S (2014) Soybean the wonder legume. Beverage Food World 31(1):61–62
Talukdar A, Shivakumar M, Verma K, Kumar A, Mukherjee K, Lal SK (2014) Genetic elimination of Kunitz trypsin inhibitor (KTI) from DS9712, an Indian soybean variety. Indian J Genet 74(4 Suppl):608–611. https://doi.org/10.5958/0975-6906.2014.00898.0
Huang YH, Ku H-M, Wang CA, Chen LY, He SS, Chen S, Liao PC, Juan PY, Kao CF (2022) A multiple phenotype imputation method for genetic diversity and core collection in Taiwanese vegetable soybean. Front Plant Sci. https://doi.org/10.3389/fpls.2022.948349
Liu C, Wang X, Chen H, Xia H, Tu B, Li Y, Zhang Q, Liu X (2022) Nutritional quality of different potassium efficiency types of vegetable soybean as affected by potassium nutrition. Food Qual Saf 1:6. https://doi.org/10.1093/fqsafe/fyab039
Liu N, Niu Y, Zhang G, Feng Z, Bo Y, Lian J, Wang B, Gong Y (2022) Genome sequencing and population resequencing provide insights into the genetic basis of domestication and diversity of vegetable soybean. Hortic Res. https://doi.org/10.1093/hr/uhab052
Saldivar XE, Wang YJ, Chen P, Mauromoustakos A (2010) Effects of blanching and storage conditions on soluble sugar contents in vegetable soybean. LWT-Food Sci Technol. https://doi.org/10.1016/j.lwt.2010.04.017
Shanmugasundaram S, Yan MR (2010) 19. Vegetable soybean. In: Singh Guriqbal Singh Guriqbal (ed) The soybean: Botany, production and uses. CABI, Wallingford, pp 427–460
Keatinge JD, Easdown WJ, Yang RY, Chadha ML, Shanmugasundaram S (2011) Overcoming chronic malnutrition in a future warming world: the key importance of mungbean and vegetable soybean. Euphytica 180(1):129–141. https://doi.org/10.1007/s10681-011-0401-6
Zhang GW, Xu SC, Mao WH, Hu QZ, Gong YM (2013) Determination of the genetic diversity of vegetable soybean [Glycine max (L.) Merr.] using EST-SSR markers. J Zhejiang Uni Sci B 14(4):279–288. https://doi.org/10.1631/jzus.B1200243
Ravishankar M, Pan RS, Kaur DP, Giri RR, Anil Kumar V, Rathore A, Easdown W, Nair RM (2016) Vegetable soybean: a crop with immense potential to improve human nutrition and diversify cropping systems in Eastern India-A Review. Soybean Res 14(2):1–13
Doldi ML, Vollmann J, Lelley T (1997) Genetic diversity in soybean as determined by RAPD and microsatellite analysis. Plant breed 116(4):331–335
Yu X, Fu X, Yang Q, Jin H, Zhu L, Yuan F (2022) Genome-wide variation analysis of four vegetable soybean cultivars based on re-sequencing. Plants (Basel) 11(1):28. https://doi.org/10.3390/plants11010028
Jankauskiene J, Brazaityte A, Vastakaite-Kairiene V (2021) Potential of vegetable soybean cultivation in Lithuania. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. https://doi.org/10.15835/nbha49112267
Fufa HU, Baenziger PS, Beecher BS, Dweikat I, Graybosch RA, Eskridge KM (2005) Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars. Euphytica 145(1):133–346. https://doi.org/10.1007/s10681-005-0626-3
Song QJ, Quigley CV, Nelson RL, Carter TE, Boerma HR, Strachan JL, Cregan PBA (1999) A selected set of trinucleotide simple sequence repeat markers for soybean cultivar identification. Plant Varieties and Seeds 12:207–220
Mimura M, Coyne CJ, Bambuck MW, Lumpkin TA (2007) SSR diversity of vegetable soybean [Glycine max (L.) Merr.]. Genetic Resour Crop Evol 54(3):497–508. https://doi.org/10.1155/2014/968796
Devi EL, Hossain F, Muthusamy V, Chhabra R, Zunjare RU, Baveja A, Jaiswal SK, Goswami R, Dosad S (2017) Microsatellite marker-based characterization of waxy maize inbreds for their utilization in hybrid breeding. 3Biotech. https://doi.org/10.1007/s13205-017-0946-8
Venison EP, Litthauer S, Laws P, Denance C, Fernandez-Fernandez F, Durel CE, Ordidge M (2022) Microsatellite markers as a tool for active germplasm management and bridging the gap between national and local collections of apples. Genetic Resour Crop Evol 69(5):1817–1832. https://doi.org/10.1007/s10722-022-01342-5
Wang L, Guan R, Zhangxiong L, Chang R, Qiu L (2006) Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci 46:1032–1038
Khanande AS, Jadhav PV, Kale PB, Madavi SM, Mohari MP, Nichal SS (2016) Genetic diversity in vegetable and grain type soybean genotypes identified using morphological descriptor and EST-SSR markers. Vegetos-An Int J Plant Res 29(3):158–173. https://doi.org/10.4172/2229-4473.1000156
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal biochem 72(1–2):248–254
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1(4):19–21
Xu SB, Tao YF, Yang ZQ, Chu JY (2002) A simple and rapid methods used for silver staining and gel preservation. Yi Chuan 24(3):335–336
Raj P, Rumit P, Dinesh P (2020) Variability: genetic variability analysis for plant breeding research. R package version 0.1. 0.
Panse VG, Sukhatme PV (1957) Genetics of quantitative characters in relation to plant breeding. Indian J Genet 17(2):318–328
Liu K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Science Publishers Montpellier, Enfield, pp 43–76
Shilpashree N, Devi SN, Manjunathagowda DC, Muddappa A, Abdelmohsen SA, Tamam N, Elansary HO, El-Abedin TK, Abdelbacki AM, Janhavi V (2021) Morphological characterization, variability and diversity among vegetable soybean (Glycine max L.) genotypes. Plants. https://doi.org/10.3390/plants10040671
Kundi RS, Gill MS, Singh TP, Phul PS (1997) Radiation induced variability for quantitative traits in soybean (Glycine max (L.) Merrill). Crop Improv 24(2):231–234
Husain SM, Bhatnagar PS, Karmakar PG (1998) Radiation induced variability for seed longevity in soybean variety NRC-7. Soybean Genetics Newsl 25:83–85
Malek MA, Rafii MY, Afroz SS, Nath UK, Mondal M (2014) Morphological characterization and assessment of genetic variability, character association, and divergence in soybean mutants. Sci World J. https://doi.org/10.1155/2014/968796
Stuti M, Pancheshwar DK, Prateek S, Avinash J (2015) Study of genetic variability in recently evolved genotypes of soybean [Glycine max (L Merill]. Biosci. Trends 8(19):5390–5393
Dong D, Fu X, Yuan F, Chen P, Zhu S, Li B, Yang Q, Yu X, Zhu D (2014) Genetic diversity and population structure of vegetable soybean (Glycine max (L.) Merr.) in China as revealed by SSR markers. Genet Resour Crop Evol. https://doi.org/10.1007/s10722-013-0024-y
Dayaman V, Senthil N, Raveendran M, Nadarajan N, Shanmugasundaram P, Balasubramanian P (2009) Diversity analysis in selected Indian soybean [Glycine max (L.) Merrill] using morphological and SSR markers. Int J Integr Biol 5(2):125–129
Kumar SP, Susmita C, Sripathy KV, Agarwal DK, Pal G, Singh AN, Kumar S, Rai AK, Simal-Gandara J (2022) Molecular characterization and genetic diversity studies of Indian soybean (Glycine max (L.) Mer.r) cultivars using SSR markers. Mol Biol Rep. 49(3):2129–2140. https://doi.org/10.1007/s11033-021-07030-4
Denwar NN, Awuku FJ, Diers B, Addae-Frimpomaah F, Chigeza G, Oteng-Frimpong R, Puozaa DK, Barnor MT (2009) Genetic diversity, population structure and key phenotypic traits driving variation within soyabean (Glycine max) collection in Ghana. Plant Breed 138(5):577–587. https://doi.org/10.1111/pbr.12700
Wang L, Guan R, Zhang Xiong L, Chang R, Qiu L (2006) Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci 46(3):1032–1038. https://doi.org/10.2135/cropsci2005.0051
Hirota T, Sayama T, Yamasaki M, Sasama H, Sugimoto T, Ishimoto M, Yoshida S (2012) Diversity and population structure of black soybean landraces originating from Tanba and neighboring regions. Breed Sci 61(5):593–601. https://doi.org/10.1270/jsbbs.61.593
Bisen A, Khare D, Nair P, Tripathi N (2014) SSR analysis of 38 genotypes of soybean (Glycine Max (L.) Merr.) genetic diversity in India. Physiol Mol Biol Plants. 21(1):109–115. https://doi.org/10.1007/s12298-014-0269-8
Ghosh J, Ghosh PD, Choudhury PR (2014) An assessment of genetic relatedness between soybean [Glycine max (L.) Merrill] cultivars using SSR markers. Am J Plant Sci. https://doi.org/10.4236/ajps.2014.520325
Hipparagi Y, Singh R, Choudhury DR, Gupta V (2017) Genetic diversity and population structure analysis of Kala bhat (Glycine max (L.) Merrill) genotypes using SSR markers. Hereditas. https://doi.org/10.1186/s41065-017-0030-8
Acknowledgements
Authors sincerely acknowledge the support received from the Head of the Centre of Excellence in Plant Biotechnology and Oilseeds Research Unit, Dr. PDKV, Akola for conduct of the experiments.
Funding
The authors declare that no funds, grants, or other support were received during the research and the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization of research (PJ & SS); Designing of the experiments (PP, PJ & SS); Contribution of experimental materials (PV); Execution of field/lab experiments and data collection (PP, DR & PS); Analysis of data and interpretation (PP, RS & DR); Analysis of SSR data and interpretation (RZ, PP); Preparation of the manuscript (PP, PJ & SS).
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pardeshi, P., Jadhav, P., Sakhare, S. et al. Morphological and microsatellite marker-based characterization and diversity analysis of novel vegetable soybean [Glycine max (L.) Merrill]. Mol Biol Rep 50, 4049–4060 (2023). https://doi.org/10.1007/s11033-023-08328-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08328-1