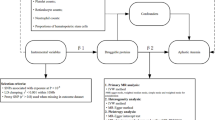
Abstract
The extraction of genomic DNA is the crucial first step in large-scale epidemiological studies. Though there are many popular DNA isolation methods from human whole blood, only a few reports have compared their efficiencies using both end-point and real-time PCR assays. Genomic DNA was extracted from coronary artery disease patients using solution-based conventional protocols such as the phenol–chloroform/proteinase-K method and a non-phenolic non-enzymatic Rapid-Method, which were evaluated and compared vis-a-vis a commercially available silica column-based Blood DNA isolation kit. The appropriate method for efficiently extracting relatively pure DNA was assessed based on the total DNA yield, concentration, purity ratios (A260/A280 and A260/A230), spectral profile and agarose gel electrophoresis analysis. The quality of the isolated DNA was further analysed for PCR inhibition using a murine specific ATP1A3 qPCR assay and mtDNA/Y-chromosome ratio determination assay. The suitability of the extracted DNA for downstream applications such as end-point SNP genotyping, was tested using PCR-RFLP analysis of the AGTR1-1166A>C variant, a mirSNP having pharmacogenetic relevance in cardiovascular diseases. Compared to the traditional phenol–chloroform/proteinase-K method, our results indicated the Rapid-Method to be a more suitable protocol for genomic DNA extraction from human whole blood in terms of DNA quantity, quality, safety, processing time and cost. The Rapid-Method, which is based on a simple salting-out procedure, is not only safe and cost-effective, but also has the added advantage of being scaled up to process variable sample volumes, thus enabling it to be applied in large-scale epidemiological studies.


Similar content being viewed by others
References
Philibert RA, Zadorozhnyaya O, Beach SR, Brody GH (2008) Comparison of the genotyping results using DNA obtained from blood and saliva. Psychiatr Genet 18:275–281
Abraham JE, Maranian MJ, Spiteri I, Russell R, Ingle S, Luccarini C, Earl HM, Pharoah PP, Dunning AM, Caldas C (2012) Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics 30:5–19
Herráez DL, Stoneking M (2008) High fractions of exogenous DNA in human buccal samples reduce the quality of large-scale genotyping. Anal Biochem 383:329–331
Godderis L, Schouteden C, Tabish A, Poels K, Hoet P, Baccarelli AA, Van Landuyt K (2015) Global methylation and hydroxymethylation in DNA from blood and saliva in healthy volunteers. Biomed Res Int 2015:845041
Al-Yatama MK, Mustafa AS, Ali S, Abraham S, Khan Z, Khaja N (2001) Detection of Y chromosome-specific DNA in the plasma and urine of pregnant women using nested polymerase chain reaction. Prenat Diagn 21:399–402
Utting M, Werner W, Dahse R, Schubert J, Junker K (2002) Microsatellite analysis of free tumor DNA in urine, serum, and plasma of patients: a minimally invasive method for the detection of bladder cancer. Clin Cancer Res 8:35–40
Bryzgunova OE, Morozkin ES, Yarmoschuk SV, Vlassov VV, Laktionov PP (2008) Methylation-specific sequencing of GSTP1 gene promoter in circulating/extracellular DNA from blood and urine of healthy donors and prostate cancer patients. Ann N Y Acad Sci 1137:222–225
Su YH, Wang M, Brenner DE, Norton PA, Block TM (2008) Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann N Y Acad Sci 1137:197–206
El Bali L, Diman A, Bernard A, Roosens NH, De Keersmaecker SC (2014) Comparative study of seven commercial kits for human DNA extraction from urine samples suitable for DNA biomarker-based public health studies. J Biomol Tech 25:96–110
Amory S, Keyser C, Crubézy E, Ludes B (2007) STR typing of ancient DNA extracted from hair shafts of Siberian mummies. Forensic Sci Int 166:218–229
Grody WW, Nakamura RM, Kiechle FL, Strom C (2009) Molecular diagnostics: techniques and applications for the clinical laboratory. Academic Press, Cambridge, p 39
Kang M-J, Yu H, Kim S-K, Park S-R, Yang I (2011) Quantification of trace-level DNA by real-time whole genome amplification. PLoS One 6:e28661
Chacon-Cortes D, Haupt L, Lea R, Griffiths L (2012) Comparison of genomic DNA extraction techniques from whole blood samples: a time, cost and quality evaluation study. Mol Biol Rep 39:5961–5966
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 1st edn. Cold Spring Harbor Laboratory Press, New York
Maloney B, Ray B, Hayden EP, Nurnberger JI Jr, Lahiri DK (2009) Development and validation of the high-quality ‘rapid method for swab’ to genotype the HTTLPR serotonin transporter (SLC6A4) promoter polymorphism. Psychiatr Genet 19:72–82
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Johar K, Priya A, Wong-Riley MT (2014) Regulation of Na(+)/K(+)-ATPase by neuron-specific transcription factor Sp4: implication in the tight coupling of energy production, neuronal activity and energy consumption in neurons. Eur J Neurosci 39:566–578
Bustin SA, Nolan T (2004) Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 15:155–166
King C, Debruyne R, Kuch M, Schwarz C, Poinar H (2009) A quantitative approach to detect and overcome PCR inhibition in ancient DNA extracts. Biotechniques 47:941–949
Viltrop T, Krjutskov K, Palta P, Metspalu A (2010) Comparison of DNA extraction methods for multiplex polymerase chain reaction. Anal Biochem 15:260–262
Lahiri DK, Nurnberger JI Jr (1991) A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res 19:5444
Lahiri DK, Bye S, Nurnberger JI Jr, Hodes ME, Crisp M (1992) A non-organic and non-enzymatic extraction method gives higher yields of genomic DNA from whole-blood samples than do nine other methods tested. J Biochem Biophys Methods 25:193–205
Opel KL, Chung D, McCord BR (2010) A study of PCR inhibition mechanisms using real time PCR. J Forensic Sci 55:25–33
Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH (2009) DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion 9:261–265
Berger A, Bruschek M, Grethen C, Sperl W, Kofler B (2001) Poor storage and handling of tissue mimics mitochondrial DNA depletion. Diagn Mol Pathol 10:55–59
Schünemann HJ, Stanulla M, Trevisan M, Aplan PD, Freudenheim JL, Muti P (2000) Short-term storage of blood samples and DNA isolation in serum separator tubes for application in epidemiological studies and clinical research. Ann Epidemiol 10:538–544
Ng DP, Koh D, Choo SG, Ng V, Fu Q (2004) Effect of storage conditions on the extraction of PCR-quality genomic DNA from saliva. Clin Chim Acta 343:191–194
Xu R, Ye P, Luo L, Wu H, Dong J, Deng X (2010) A simple and efficient method for DNA purification from samples of highly clotted blood. Mol Biotechnol 46:258–264
Choi EH, Lee SK, Ihm C, Sohn YH (2014) Rapid DNA extraction from dried blood spots on filter paper: potential applications in biobanking. Osong Public Health Res Perspect 5:351–357
Clements DN, Wood S, Carter SD, Ollier WE (2008) Assessment of the quality and quantity of genomic DNA recovered from canine blood samples by three different extraction methods. Res Vet Sci 85:74–79
Psifidi A, Dovas CI, Bramis G, Lazou T, Russel CL, Arsenos G, Banos G (2015) Comparison of eleven methods for genomic DNA extraction suitable for large-scale whole-genome genotyping and long-term DNA banking using blood samples. PLoS One 10:e0115960
Acknowledgement
LK acknowledges the Science and Engineering Research Board-Department of Science and Technology (SERB-DST), Govt. of India, New Delhi, for financial support via the Fast Track Young Scientists Scheme, Project File No. SB/YS/LS-174/2013. We thank Dr. Mahesh S. K. for kindly providing mouse cDNA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest (financial or non-financial).
Informed consent
Written informed consent was obtained from all participants.
Research involving human participants
The study protocol was approved by the Institutional Ethics Committee for Clinical Studies at the SCTIMST, IEC No. SCT/IEC/558/JUNE-2014.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koshy, L., Anju, A.L., Harikrishnan, S. et al. Evaluating genomic DNA extraction methods from human whole blood using endpoint and real-time PCR assays. Mol Biol Rep 44, 97–108 (2017). https://doi.org/10.1007/s11033-016-4085-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-4085-9