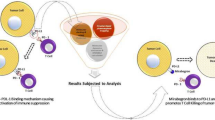
Abstract
Endometrial cancer (EC) is the 6th most common cancer in women around the world. Alone in the United States (US), 66,200 new cases and 13,030 deaths are expected to occur in 2023 which needs the rapid development of potential therapies against EC. Here, a network-based drug-repurposing strategy is developed which led to the identification of 16 FDA-approved drugs potentially repurposable for EC as potential immune checkpoint inhibitors (ICIs). A network of EC-associated immune checkpoint proteins (ICPs)-induced protein interactions (P-ICP) was constructed. As a result of network analysis of P-ICP, top key target genes closely interacting with ICPs were shortlisted followed by network proximity analysis in drug–target interaction (DTI) network and pathway cross-examination which identified 115 distinct pathways of approved drugs as potential immune checkpoint inhibitors. The presented approach predicted 16 drugs to target EC-associated ICPs-induced pathways, three of which have already been used for EC and six of them possess immunomodulatory properties providing evidence of the validity of the strategy. Classification of the predicted pathways indicated that 15 drugs can be divided into two distinct pathway groups, containing 17 immune pathways and 98 metabolic pathways. In addition, drug–drug correlation analysis provided insight into finding useful drug combinations. This fair and verified analysis creates new opportunities for the quick repurposing of FDA-approved medications in clinical trials.
Graphical abstract







Similar content being viewed by others
Data availability
The supplementary data Tables and figures are provided.
Code availability
The coding files will be provided on demand.
References
Rodrigues R, Duarte D, Vale N (2022) Drug repurposing in cancer therapy: influence of patient’s genetic background in breast cancer treatment. Int J Mol Sci. https://doi.org/10.3390/IJMS23084280
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108. https://doi.org/10.3322/CAAC.21262
Zou Y, Xu Y, Chen X, Zheng L (2023) Advances in the application of immune checkpoint inhibitors in gynecological tumors. Int Immunopharmacol 117:109774. https://doi.org/10.1016/j.intimp.2023.109774
Lin CK, Liu ST, Wu ZS, Wang YC, Huang SM (2021) Mechanisms of cisplatin in combination with repurposed drugs against human endometrial carcinoma cells. Life (Basel, Switzerland) 11(2):1–18. https://doi.org/10.3390/LIFE11020160
Gómez-Raposo C, Merino Salvador M, Aguayo Zamora C, García de Santiago B, Casado Sáenz E (2021) Immune checkpoint inhibitors in endometrial cancer. Crit Rev Oncol Hematol 161:103306. https://doi.org/10.1016/J.CRITREVONC.2021.103306
Ahmed F et al (2022) A comprehensive review of artificial intelligence and network based approaches to drug repurposing in Covid-19. Biomed Pharmacother 153:113350. https://doi.org/10.1016/J.BIOPHA.2022.113350
Ahmed F, Samantasinghar A, Soomro AM (2023) A systematic review of computational approaches to understand cancer biology for informed drug repurposing. J Biomed Inform. https://doi.org/10.1016/j.jbi.2023.104373
Ahmed F et al (2023) Drug repurposing for viral cancers: a paradigm of machine learning, deep learning, and virtual screening-based approaches. J Med Virol. https://doi.org/10.1002/jmv.28693
Samantasinghar A et al (2023) A comprehensive review of key factors affecting the efficacy of antibody drug conjugate. Biomed Pharmacother 161:114408. https://doi.org/10.1016/j.biopha.2023.114408
Asif A et al (2021) Microphysiological system with continuous analysis of albumin for hepatotoxicity modeling and drug screening. J Ind Eng Chem 98:318–326. https://doi.org/10.1016/J.JIEC.2021.03.035
Ahmed F et al (2022) Drug repurposing in psoriasis, performed by reversal of disease-associated gene expression profiles. Comput Struct Biotechnol J 20:6097–6107. https://doi.org/10.1016/J.CSBJ.2022.10.046
Ahmed F et al (2022) SperoPredictor: an integrated machine learning and molecular docking-based drug repurposing framework with use case of COVID-19. Front Public Health. https://doi.org/10.3389/FPUBH.2022.902123
Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6(1):1–18. https://doi.org/10.1038/s41421-020-0153-3
Shiravand Y et al (2022) Immune checkpoint inhibitors in cancer therapy. Curr Oncol 29(5):3044–3060. https://doi.org/10.3390/curroncol29050247
Hirschhorn D et al (2023) T cell immunotherapies engage neutrophils to eliminate tumor antigen escape variants. Cell 186(7):1432-1447.e17. https://doi.org/10.1016/j.cell.2023.03.007
Sharma R, Kaur G, Bansal P, Chawla V, Gupta V (2022) Bioinformatics paradigms in drug discovery and drug development. Curr Top Med Chem 23(7):579–588. https://doi.org/10.2174/1568026623666221229113456
Maiorano BA, Maiorano MFP, Cormio G, Maglione A, Lorusso D, Maiello E (2022) How immunotherapy modified the therapeutic scenario of endometrial cancer: a systematic review. Front Oncol. https://doi.org/10.3389/fonc.2022.844801
Musacchio L et al (2020) Immune checkpoint inhibitors: a promising choice for endometrial cancer patients? J Clin Med 9(6):1721. https://doi.org/10.3390/JCM9061721
Napolitano F et al (2013) Drug repositioning: a machine-learning approach through data integration. J Cheminform 5(6):1–9. https://doi.org/10.1186/1758-2946-5-30/FIGURES/5
Yuan M et al (2022) A gene co-expression network-based drug repositioning approach identifies candidates for treatment of hepatocellular carcinoma. Cancers (Basel) 14(6):1573. https://doi.org/10.3390/CANCERS14061573/S1
Zhou Y, Wang F, Tang J, Nussinov R, Cheng F (2020) Artificial intelligence in COVID-19 drug repurposing. Lancet Digit Health 2(12):e667–e676. https://doi.org/10.1016/S2589-7500(20)30192-8
Yang S, Fu C, Lian X, Dong X, Zhang Z (2019) Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. mSystems. https://doi.org/10.1128/MSYSTEMS.00303-18/SUPPL_FILE/MSYSTEMS.00303-18-SF006.PDF
Liu C et al (2020) Computational network biology: data, models, and applications. Phys Rep 846:1–66. https://doi.org/10.1016/J.PHYSREP.2019.12.004
Firoozbakht F, Rezaeian I, Rueda L, Ngom A (2022) Computationally repurposing drugs for breast cancer subtypes using a network-based approach. BMC Bioinform 23(1):1–36. https://doi.org/10.1186/S12859-022-04662-6/TABLES/2
Vitali F et al (2016) A network-based data integration approach to support drug repurposing and multi-target therapies in triple negative breast cancer. PLoS ONE 11(9):e0162407. https://doi.org/10.1371/JOURNAL.PONE.0162407
Fang J et al (2020) Network-based translation of GWAS findings to pathobiology and drug repurposing for Alzheimer’s disease. medRxiv. https://doi.org/10.1101/2020.01.15.20017160
Cheng F et al (2018) Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun 9(1):1–12. https://doi.org/10.1038/s41467-018-05116-5
Cheng F et al (2019) A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat Commun 10(1):1–14. https://doi.org/10.1038/s41467-019-10744-6
Wang Y et al (2022) Immune checkpoint modulators in cancer immunotherapy: recent advances and emerging concepts. J Hematol Oncol 15(1):1–53. https://doi.org/10.1186/S13045-022-01325-0
Vafaei S et al (2022) Combination therapy with immune checkpoint inhibitors (ICIs); a new frontier. Cancer Cell Int 22(1):1–27. https://doi.org/10.1186/s12935-021-02407-8
Naimi A et al (2022) Tumor immunotherapies by immune checkpoint inhibitors (ICIs); the pros and cons. Cell Commun Signal 20(1):1–31. https://doi.org/10.1186/S12964-022-00854-Y
Khalifa R, Elsese N, El-Desouky K, Shaair H, Helal D (2021) Immune checkpoint proteins (PD-L1 and CTLA-4) in endometrial carcinoma: prognostic role and correlation with CD4+/CD8+ tumor infiltrating lymphocytes (TILs) ratio. J Immunoassay Immunochem 43(2):192–212. https://doi.org/10.1080/15321819.2021.1981377
Hagberg A, Swart P, Schult J, Daniel A (2008) Exploring network structure, dynamics, and function using networkx (conference) | OSTI.GOV. https://www.osti.gov/biblio/960616. Colgate University, Hamilton, NY (United States), Accessed 15 Mar 2023
Szklarczyk D et al (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49(D1):D605–D612. https://doi.org/10.1093/nar/gkaa1074
Valdeolivas A et al (2019) Random walk with restart on multiplex and heterogeneous biological networks. Bioinformatics 35(3):497–505. https://doi.org/10.1093/bioinformatics/bty637
Newman MEJ (2003) The structure and function of complex networks. SIAM Rev 45(2):167–256. https://doi.org/10.1137/s003614450342480
Ahmed F et al (2022) Decade of bio-inspired soft robots: a review. Smart Mater Struct 31(7):073002. https://doi.org/10.1088/1361-665X/AC6E15
Han N et al (2021) Identification of SARS-CoV-2–induced pathways reveals drug repurposing strategies. Sci Adv. https://doi.org/10.1126/SCIADV.ABH3032/SUPPL_FILE/ABH3032_TABLE_S9.XLSX
Xie Z et al (2021) Gene set knowledge discovery with Enrichr. Curr Protoc 1(3):e90. https://doi.org/10.1002/CPZ1.90
Huang DW et al (2007) The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8(9):1–16. https://doi.org/10.1186/GB-2007-8-9-R183/TABLES/3
Wishart DS et al (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34(Database issue):D668. https://doi.org/10.1093/NAR/GKJ067
Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P (2008) STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. https://doi.org/10.1093/nar/gkm795
Cheng F, István I, Kovácskovács A, László A-L, Lászlóbarabási L (2019) Network-based prediction of drug combinations. Nat Commun 10(1):1–11. https://doi.org/10.1038/s41467-019-09186-x
Ahmed F et al (2022) Multi-material bio-inspired soft octopus robot for underwater synchronous swimming. J Bion Eng. https://doi.org/10.1007/S42235-022-00208-X
Guney E, Menche J, Vidal M, Barábasi AL (2016) Network-based in silico drug efficacy screening. Nat Commun 7(May 2015):1–13. https://doi.org/10.1038/ncomms10331
Wu Z, Peng Y, Yu Z, Li W, Liu G, Tang Y (2020) NetInfer: a web server for prediction of targets and therapeutic and adverse effects via network-based inference methods. J Chem Inf Model 60(8):3687–3691. https://doi.org/10.1021/ACS.JCIM.0C00291/SUPPL_FILE/CI0C00291_SI_002.XLSX
Oh MS, Chae YK (2019) Deep and durable response with combination CTLA-4 and PD-1 blockade in mismatch repair (MMR)-proficient endometrial cancer. J Immunother 42(2):51–54. https://doi.org/10.1097/CJI.0000000000000244
Friedman LA, Ring KL, Mills AM (2020) LAG-3 and GAL-3 in endometrial carcinoma: emerging candidates for immunotherapy. Int J Gynecol Pathol 39(3):203–212. https://doi.org/10.1097/PGP.0000000000000608
Singh MM, Singh E, Miller H, Strum WB, Coyle W (2012) Colorectal cancer screening in women with endometrial cancer: are we following the guidelines? J Gastrointest Cancer 43(2):190–195. https://doi.org/10.1007/S12029-011-9271-3
Visnyei K, Shahrokni A, Hashmi S, Orell J, Wild DMG (2012) A case of groans, moans and stones with malignant undertones: endometrioid carcinoma-associated hypercalcemia. Oncol Lett 3(2):335. https://doi.org/10.3892/OL.2011.475
Soisson S et al (2018) Long-term cardiovascular outcomes among endometrial cancer survivors in a large, population-based cohort study. J Natl Cancer Inst 110(12):1342. https://doi.org/10.1093/JNCI/DJY070
Jin J, Dalwadi SM, Masand RP, Hall TR, Anderson ML, Ludwig MS (2020) Association between metabolic syndrome and endometrial cancer survival in a SEER-Medicare linked database. Am J Clin Oncol. https://doi.org/10.1097/COC.0000000000000686
Leskovec J, Krevl A (2014) Stanford large network dataset collection. https://snap.stanford.edu/data/. Accessed 15 Mar 2023
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. In: BT—international AAAI conference on weblogs and social. pp. 361–362
Gaulton A et al (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40(Database issue):D1100. https://doi.org/10.1093/NAR/GKR777
Preto AJ, Matos-Filipe P, Mourão J, Moreira IS (2022) SYNPRED: prediction of drug combination effects in cancer using different synergy metrics and ensemble learning. GigaScience 11:1–15. https://doi.org/10.1093/GIGASCIENCE/GIAC087
Foucquier J, Guedj M (2015) Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect 3(3):e00149. https://doi.org/10.1002/PRP2.149
Ianevski A, Timonen S, Kononov A, Aittokallio T, Giri AK (2020) SynToxProfiler: an interactive analysis of drug combination synergy, toxicity and efficacy. PLoS Comput Biol 16(2):e1007604. https://doi.org/10.1371/JOURNAL.PCBI.1007604
Lehár J et al (2009) Synergistic drug combinations improve therapeutic selectivity. Nat Biotechnol 27(7):659. https://doi.org/10.1038/NBT.1549
Duarte D, Vale N (2022) Evaluation of synergism in drug combinations and reference models for future orientations in oncology. Curr Res Pharmacol Drug Discov 3:100110. https://doi.org/10.1016/J.CRPHAR.2022.100110
Chen H, Zhang Z, Zhang J (2022) In silico drug repositioning based on integrated drug targets and canonical correlation analysis. BMC Med Genomics. https://doi.org/10.1186/S12920-022-01203-1
Luo S, Lin R, Liao X, Li D, Qin Y (2021) Identification and verification of the molecular mechanisms and prognostic values of the cadherin gene family in gastric cancer. Sci Rep 11(1):23674. https://doi.org/10.1038/S41598-021-03086-1
Brasky TM et al (2017) Nonsteroidal anti-inflammatory drugs and endometrial carcinoma mortality and recurrence. J Natl Cancer Inst. https://doi.org/10.1093/JNCI/DJW251
Ando H et al (2017) Panobinostat enhances growth suppressive effects of progestin on endometrial carcinoma by increasing progesterone receptor and mitogen-inducible gene-6. Horm Cancer 8(4):257–267. https://doi.org/10.1007/S12672-017-0295-4
Audrito V (2021) The extracellular NADome modulates immune responses. Front Immunol 12(August):17–20. https://doi.org/10.3389/fimmu.2021.704779
Omran HM, Almaliki MS (2020) Influence of NAD+ as an ageing-related immunomodulator on COVID 19 infection: a hypothesis. J Infect Public Health 13(9):1196. https://doi.org/10.1016/J.JIPH.2020.06.004
Xu X, Li X, Zhao Y, Huang H (2021) Immunomodulatory effects of histone deacetylation inhibitors in graft-vs.-host disease after allogeneic stem cell transplantation. Front Immunol 12:481. https://doi.org/10.3389/FIMMU.2021.641910/BIBTEX
Díez-Tercero L, Delgado LM, Bosch-Rué E, Perez RA (2021) Evaluation of the immunomodulatory effects of cobalt, copper and magnesium ions in a pro inflammatory environment. Sci Rep 11(1):1–13. https://doi.org/10.1038/s41598-021-91070-0
Ashraf Z et al (2019) Dexibuprofen amide derivatives as potential anticancer agents: synthesis, in silico docking, bioevaluation, and molecular dynamic simulation. Drug Des Dev Ther 13:1643. https://doi.org/10.2147/DDDT.S178595
Yao W, Wang F, Wang H (2016) Immunomodulation of artemisinin and its derivatives. Sci Bull 61(18):1399–1406. https://doi.org/10.1007/S11434-016-1105-Z
Mallat Z, Lobo SM, Malik A, Tong S (2021) 561. Phase 3 trial of fostamatinib for the treatment of COVID-19: repurposing an immunomodulatory drug previously approved for immune thrombocytopenia. Open Forum Infect Dis 8(Suppl 1):S382. https://doi.org/10.1093/OFID/OFAB466.759
Migliorisi G et al (2022) Elexacaftor-tezacaftor-ivacaftor as a final frontier in the treatment of cystic fibrosis: definition of the clinical and microbiological implications in a case-control study. Pharmaceuticals. https://doi.org/10.3390/PH15050606
Teng F et al (2016) Treatment with an SLC12A1 antagonist inhibits tumorigenesis in a subset of hepatocellular carcinomas. Oncotarget 7(33):53571–53582. https://doi.org/10.18632/oncotarget.10670
Malebari AM et al (2020) The anticancer activity for the bumetanide-based analogs via targeting the tumor-associated membrane-bound human carbonic anhydrase-IX enzyme. Pharmaceuticals (Basel). https://doi.org/10.3390/ph13090252
Endo H, Yano M, Okumura Y, Kido H (2014) Ibuprofen enhances the anticancer activity of cisplatin in lung cancer cells by inhibiting the heat shock protein 70. Cell Death Dis 5(1):e1027. https://doi.org/10.1038/cddis.2013.550
Shen W et al (2020) Ibuprofen mediates histone modification to diminish cancer cell stemness properties via a COX2-dependent manner. Br J Cancer 123(5):730–741. https://doi.org/10.1038/s41416-020-0906-7
Helland Ø, Popa M, Bischof K, Gjertsen BT, McCormack E, Bjørge L (2016) The HDACi panobinostat shows growth inhibition both in vitro and in a bioluminescent orthotopic surgical xenograft model of ovarian cancer. PLoS ONE 11(6):e0158208. https://doi.org/10.1371/journal.pone.0158208
Garrett LA, Growdon WB, Rueda BR, Foster R (2016) Influence of a novel histone deacetylase inhibitor panobinostat (LBH589) on the growth of ovarian cancer. J Ovarian Res 9(1):58. https://doi.org/10.1186/s13048-016-0267-2
Jeske R, Yuan X, Fu Q, Bunnell BA, Logan TM, Li Y (2021) In vitro culture expansion shifts the immune phenotype of human adipose-derived mesenchymal stem cells. Front Immunol 12:621744. https://doi.org/10.3389/fimmu.2021.621744
Cheng M-H et al (2020) Prolonging the half-life of histone deacetylase inhibitor belinostat via 50 nm scale liposomal subcutaneous delivery system for peripheral T-cell lymphoma. Cancers (Basel). https://doi.org/10.3390/cancers12092558
Shen Y et al (2022) The histone deacetylase inhibitor belinostat ameliorates experimental autoimmune encephalomyelitis in mice by inhibiting TLR2/MyD88 and HDAC3/NF-κB p65-mediated neuroinflammation. Pharmacol Res 176:105969. https://doi.org/10.1016/j.phrs.2021.105969
Llopiz D et al (2019) Enhanced anti-tumor efficacy of checkpoint inhibitors in combination with the histone deacetylase inhibitor Belinostat in a murine hepatocellular carcinoma model. Cancer Immunol Immunother 68(3):379–393. https://doi.org/10.1007/s00262-018-2283-0
Cheng F et al (2022) Relationship between copper and immunity: the potential role of copper in tumor immunity. Front Oncol 12:1019153. https://doi.org/10.3389/fonc.2022.1019153
Farsam V, Hassan ZM, Zavaran-Hosseini A, Noori S, Mahdavi M, Ranjbar M (2011) Antitumor and immunomodulatory properties of artemether and its ability to reduce CD4+ CD25+ FoxP3+ T reg cells in vivo. Int Immunopharmacol 11(11):1802–1808. https://doi.org/10.1016/j.intimp.2011.07.008
Li T et al (2012) Anti-inflammatory and immunomodulatory mechanisms of artemisinin on contact hypersensitivity. Int Immunopharmacol 12(1):144–150. https://doi.org/10.1016/j.intimp.2011.11.004
Park SR et al (2013) A multi-histology trial of fostamatinib in patients with advanced colorectal, non-small cell lung, head and neck, thyroid, and renal cell carcinomas, and pheochromocytomas. Cancer Chemother Pharmacol 71(4):981–990. https://doi.org/10.1007/s00280-013-2091-3
Graeber S et al (2023) Effects of elexacaftor/tezacaftor/ivacaftor on sputum viscoelastic properties, airway infection and inflammation in patients with cystic fibrosis. Jahrestagung der Gesellschaft für Pädiatrische Pneumol. https://doi.org/10.1055/s-0043-1761569
Samantasinghar A, Ahmed F, Rahim CSA, Kim KH, Kim S, Choi KH (2023) Artificial intelligence-assisted repurposing of lubiprostone alleviates tubulointerstitial fibrosis. Transl Res. https://doi.org/10.1016/j.trsl.2023.07.010
Fiscon G, Conte F, Farina L, Paci P (2022) A comparison of network-based methods for drug repurposing along with an application to human complex diseases. Int J Mol Sci. https://doi.org/10.3390/ijms23073703
Fiscon G, Conte F, Farina L, Paci P (2021) SAveRUNNER: a network-based algorithm for drug repurposing and its application to COVID-19. PLoS Comput Biol 17(2):e1008686. https://doi.org/10.1371/JOURNAL.PCBI.1008686
Zeng X et al (2020) Target identification among known drugs by deep learning from heterogeneous networks. Chem Sci 11(7):1775–1797. https://doi.org/10.1039/c9sc04336e
Zeng X et al (2020) Network-based prediction of drug–target interactions using an arbitrary-order proximity embedded deep forest. Bioinformatics 36(9):2805–2812. https://doi.org/10.1093/BIOINFORMATICS/BTAA010
Bedi O, Dhawan V, Sharma PL, Kumar P (2016) Pleiotropic effects of statins: new therapeutic targets in drug design. Naunyn Schmiedebergs Arch Pharmacol 389(7):695–712. https://doi.org/10.1007/S00210-016-1252-4
Acknowledgements
This work was supported by 2023 Jeju National University, South Korea.
Funding
This work was supported by 2023 Jeju National University, South Korea.
Author information
Authors and Affiliations
Contributions
FA: conceptualization, formal analysis, methodology, original draft, writing—review & editing, data curation, and images. AS: conceptualization, formal analysis, writing—review & editing. WA: formal analysis, investigation, validation. KHC: conceptualization, review & editing, resources, supervision, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest of interest at this point.
Ethics approval and consent to participate
Will be provided on demand.
Consent for publication
The authors provide consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahmed, F., Samantasinghar, A., Ali, W. et al. Network-based drug repurposing identifies small molecule drugs as immune checkpoint inhibitors for endometrial cancer. Mol Divers (2024). https://doi.org/10.1007/s11030-023-10784-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10784-7