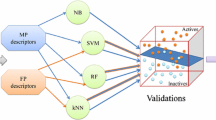
Abstract
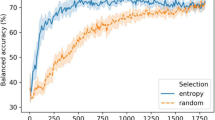
In this study, we built classification models using machine learning techniques to predict the bioactivity of non-covalent inhibitors of Bruton’s tyrosine kinase (BTK) and to provide interpretable and transparent explanations for these predictions. To achieve this, we gathered data on BTK inhibitors from the Reaxys and ChEMBL databases, removing compounds with covalent bonds and duplicates to obtain a dataset of 3895 inhibitors of non-covalent. These inhibitors were characterized using MACCS fingerprints and Morgan fingerprints, and four traditional machine learning algorithms (decision trees (DT), random forests (RF), support vector machines (SVM), and extreme gradient boosting (XGBoost)) were used to build 16 classification models. In addition, four deep learning models were developed using deep neural networks (DNN). The best model, Model D_4, which was built using XGBoost and MACCS fingerprints, achieved an accuracy of 94.1% and a Matthews correlation coefficient (MCC) of 0.75 on the test set. To provide interpretable explanations, we employed the SHAP method to decompose the predicted values into the contributions of each feature. We also used K-means dimensionality reduction and hierarchical clustering to visualize the clustering effects of molecular structures of the inhibitors. The results of this study were validated using crystal structures, and we found that the interaction between the BTK amino acid residue and the important features of clustered scaffold was consistent with the known properties of the complex crystal structures. Overall, our models demonstrated high predictive ability and a qualitative model can be converted to a quantitative model to some extent by SHAP, making them valuable for guiding the design of new BTK inhibitors with desired activity.






Similar content being viewed by others
References
Tankiewicz-Kwedlo A, Hermanowicz JM, Domaniewski T et al (2018) Simultaneous use of erythropoietin and LFM-A13 as a new therapeutic approach for colorectal cancer. Br J Pharmacol 175:743–762. https://doi.org/10.1111/bph.14099
Honigberg LA, Smith AM, Sirisawad M et al (2010) The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci USA 107:13075–13080. https://doi.org/10.1073/pnas.1004594107
Burger JA, Wiestner A (2018) Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nat Rev Cancer 18:148–167. https://doi.org/10.1038/nrc.2017.121
Li X, Zuo Y, Tang G et al (2014) Discovery of a series of 2,5-diaminopyrimidine covalent irreversible inhibitors of Bruton’s tyrosine kinase with in vivo antitumor activity. J Med Chem 57:5112–5128. https://doi.org/10.1021/jm4017762
Wu J, Zhang M, Liu D (2016) Acalabrutinib (ACP-196): a selective second-generation BTK inhibitor. J Hematol Oncol 9:21. https://doi.org/10.1186/s13045-016-0250-9
Zou Y-X, Zhu H-Y, Li X-T et al (2019) The impacts of zanubrutinib on immune cells in patients with chronic lymphocytic leukemia/small lymphocytic lymphoma. Hematol Oncol 37:392–400. https://doi.org/10.1002/hon.2667
Dhillon S (2020) Tirabrutinib: first approval. Drugs 80:835–840. https://doi.org/10.1007/s40265-020-01318-8
Dhillon S (2021) Orelabrutinib: first approval. Drugs 81:503–507. https://doi.org/10.1007/s40265-021-01482-5
Wang Y, Zhang LL, Champlin RE, Wang ML (2015) Targeting Bruton’s tyrosine kinase with ibrutinib in B-cell malignancies. Clin Pharmacol Ther 97:455–468. https://doi.org/10.1002/cpt.85
Hu N, Wang F, Sun T et al (2021) Follicular lymphoma–associated BTK mutations are inactivating resulting in augmented AKT activation. Clin Cancer Res 27:2301–2313. https://doi.org/10.1158/1078-0432.CCR-20-3741
Ma B, Bohnert T, Otipoby KL et al (2020) Discovery of BIIB068: a selective, potent, reversible Bruton’s tyrosine kinase inhibitor as an orally efficacious agent for autoimmune diseases. J Med Chem 63:12526–12541. https://doi.org/10.1021/acs.jmedchem.0c00702
Elemam NM, Hachim MY, Hannawi S, Maghazachi AA (2020) Differentially expressed genes of natural killer cells can distinguish rheumatoid arthritis patients from healthy controls. Genes 11:492. https://doi.org/10.3390/genes11050492
Voice AT, Tresadern G, Twidale RM et al (2021) Mechanism of covalent binding of ibrutinib to Bruton’s tyrosine kinase revealed by QM/MM calculations. Chem Sci 12:5511–5516. https://doi.org/10.1039/D0SC06122K
Jackson PA, Widen JC, Harki DA, Brummond KM (2017) Covalent modifiers: a chemical perspective on the reactivity of α, β-unsaturated carbonyls with thiols via hetero-michael addition reactions. J Med Chem 60:839–885. https://doi.org/10.1021/acs.jmedchem.6b00788
Crawford JJ, Johnson AR, Misner DL et al (2018) Discovery of GDC-0853: a potent, selective, and noncovalent Bruton’s tyrosine kinase inhibitor in early clinical development. J Med Chem 61:2227–2245. https://doi.org/10.1021/acs.jmedchem.7b01712
Reiff SD, Mantel R, Smith LL et al (2018) The BTK inhibitor ARQ 531 targets ibrutinib-resistant CLL and richter transformation. Cancer Discov 8:1300–1315. https://doi.org/10.1158/2159-8290.CD-17-1409
Thieme E, Liu T, Bruss N et al (2022) Dual BTK/SYK inhibition with CG-806 (luxeptinib) disrupts B-cell receptor and Bcl-2 signaling networks in mantle cell lymphoma. Cell Death Dis 13:1–11. https://doi.org/10.1038/s41419-022-04684-1
Kawahata W, Asami T, Kiyoi T et al (2021) Discovery of AS-1763: a potent, selective, noncovalent, and orally available inhibitor of Bruton’s Tyrosine Kinase. J Med Chem 64:14129–14141. https://doi.org/10.1021/acs.jmedchem.1c01279
Keam SJ (2023) Pirtobrutinib: first approval. Drugs 83:547–553. https://doi.org/10.1007/s40265-023-01860-1
Yang Z, Tian Y, Kong Y et al (2022) Classification of JAK1 inhibitors and SAR research by machine learning methods. Artif Intell Life Sci 2:100039. https://doi.org/10.1016/j.ailsci.2022.100039
Wang J, Ran T, Chen Y, Lu T (2020) Bayesian machine learning to discover Bruton’s tyrosine kinase inhibitors. Chem Biol Drug Des 96:1114–1122. https://doi.org/10.1111/cbdd.13656
Ma W, Wang Y, Chu D, Yan H (2019) 4D-QSAR and MIA-QSAR study on the Bruton’s tyrosine kinase (Btk) inhibitors. J Mol Graph Model 92:357–362. https://doi.org/10.1016/j.jmgm.2019.08.009
Márquez E, Mora JR, Flores-Morales V et al (2020) Modeling the Antileukemia activity of ellipticine-related compounds: QSAR and molecular docking study. Molecules 25:24. https://doi.org/10.3390/molecules25010024
Xing G, Liang L, Deng C et al (2020) Activity Prediction of small molecule inhibitors for antirheumatoid arthritis targets based on artificial intelligence. ACS Comb Sci 22:873–886. https://doi.org/10.1021/acscombsci.0c00169
Reaxys. https://www.reaxys.com/. Accessed 27 Dec 2022
ChEMBL Database. https://www.ebi.ac.uk/chembl/. Accessed 27 Dec 2022
Watterson SH, Tebben AJ, Ahmad S (2016) Tricyclic Atropisomer Compounds, WO2016065222A1
Hopkins BT, Bame E, Bell N et al (2019) Optimization of novel reversible Bruton’s tyrosine kinase inhibitors identified using tethering-fragment-based screens. Bioorg Med Chem 27:2905–2913. https://doi.org/10.1016/j.bmc.2019.05.021
Sutanto F, Konstantinidou M, Dömling A (2020) Covalent inhibitors: a rational approach to drug discovery. RSC Med Chem 11:876–884. https://doi.org/10.1039/D0MD00154F
Van Drie JH, Weininger D, Martin YC (1989) ALADDIN: an integrated tool for computer-assisted molecular design and pharmacophore recognition from geometric, steric, and substructure searching of three-dimensional molecular structures. J Comput-Aided Mol Des 3:225–251. https://doi.org/10.1007/BF01533070
sonnia. https://mn-am.com/products/sonnia. Accessed 27 Dec 2022
Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50:742–754. https://doi.org/10.1021/ci100050t
scikit-learn. https://scikit-learn.org/stable/. Accessed 30 Apr 2023
XGBoost. https://xgboost.readthedocs.io/en/stable/. Accessed 30 Apr 2023
Chen Y, Huang W, Nguyen L, Weng T-W (2021) On the Equivalence between Neural Network and Support Vector Machine. In: Advances in Neural Information Processing Systems. Curran Associates, Inc., pp 23478–23490
PyTorch. https://www.pytorch.org. Accessed 30 Apr 2023
Louppe G (2014) Understanding random forests: from theory to practice. Preprint at http://arxiv.org/14077502
Ribeiro MT, Singh S, Guestrin C (2016) “Why should i trust you?” Explaining the predictions of any classifier. in: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. pp 1135–1144
Lundberg SM, Lee S-I (2017) A unified approach to interpreting model predictions. Adv Neural Inf Process Syst 30:4768–4777
Chaudhuri D, Chaudhuri BB (1997) A novel multiseed nonhierarchical data clustering technique. IEEE Trans on Syst, Man, Cybern Part B (Cybernetics) 27:871–876. https://doi.org/10.1109/3477.623240
van der Maaten LJP, Hinton GE (2008) Visualizing high-dimensional data using t-SNE. J Mach Learn Res 9:2579–2605
Gomez EB, Ebata K, Randeria HS et al (2023) Pirtobrutinib preclinical characterization: a highly selective, non-covalent (reversible) BTK inhibitor. Blood. https://doi.org/10.1182/blood.2022018674
Watterson SH, De Lucca GV, Shi Q et al (2016) Discovery of 6-fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide (BMS-986142): a reversible inhibitor of Bruton’s Tyrosine Kinase (BTK) conformationally constrained by two locked atropisomers. J Med Chem 59:9173–9200. https://doi.org/10.1021/acs.jmedchem.6b01088
Liu J, Guiadeen D, Krikorian A et al (2016) Discovery of 8-amino-imidazo[1,5- a ]pyrazines as reversible BTK Inhibitors for the treatment of rheumatoid arthritis. ACS Med Chem Lett 7:198–203. https://doi.org/10.1021/acsmedchemlett.5b00463
Liu J, Guiadeen D, Krikorian A et al (2020) Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes. Bioorg Med Chem Lett 30:127390. https://doi.org/10.1016/j.bmcl.2020.127390
Hopkins BT, Bame E, Bajrami B et al (2022) Discovery and preclinical characterization of BIIB091, a reversible, selective BTK inhibitor for the treatment of multiple sclerosis. J Med Chem 65:1206–1224. https://doi.org/10.1021/acs.jmedchem.1c00926
Qiu H, Ali Z, Bender A et al (2021) Discovery of potent and selective reversible Bruton’s tyrosine kinase inhibitors. Bioorg Med Chem 40:116163. https://doi.org/10.1016/j.bmc.2021.116163
Acknowledgements
We thank the Molecular Networks GmbH, Nuremberg, Germany for providing the programs SONNIA for our scientific work.
Author information
Authors and Affiliations
Contributions
Data collection, main modeling, and manuscript drafting: GL; k-means algorithm script, article grammar modification, and guidance: JL and YT; DNN algorithm script, submission guidance, and submission format modification: YZ; Supplemental calculation of the latest crystal structure 8FLL: XP; Correspondence: AY. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, G., Li, J., Tian, Y. et al. Machine learning-based classification models for non-covalent Bruton’s tyrosine kinase inhibitors: predictive ability and interpretability. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10696-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10696-6