Abstract
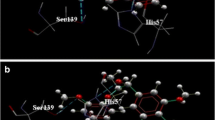
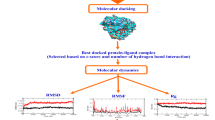
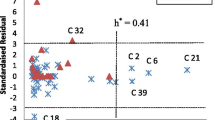
Hepatitis C virus, a member of the Flaviviridae family and genus Hepacivirus, is an enveloped, positively single stranded RNA virus. Its surface consists of a heterodimer of E1 and E2 proteins which play a crucial role in receptor binding and membrane fusion. In this study we have used in silico virtual screening by utilizing ensemble docking on the approved drugs. These drugs can bind with high efficiency to the 36 prominent conformations of the CD81 binding site clustered from a total of 3 µs MD simulation data on the E2 protein. We started with 9213 compounds from the FDA list of drugs and progressively came down to 5 compounds which have been seen to bind with very high efficiency to not only all the conformations but also the two predicted druggable pockets that encompass the CD81 binding site. MM/PBSA binding energy calculations also point to the highly stable interaction of the compounds to the E2 protein. This study may in future broaden the arsenal of therapeutics for use against HCV infection and lead to more effective care against the virus.











Similar content being viewed by others
References
Lindenbach BD, Rice CM (2005) Unravelling hepatitis C virus replication from genome to function. Nature 436(7053):933–938
Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA et al (1999) The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med 341(8):556–562
Kato N (2000) Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microb Comp Genomics 5(3):129–151
Zein NN (2000) Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev 13(2):223–235. https://doi.org/10.1128/CMR.13.2.223
Bukh J, Miller RH, Purcell RH (1995) Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis 15:41–63
https://www.who.int/news-room/fact-sheets/detail/hepatitis-c
Spearman CW, Dusheiko GM, Hellard M, Sonderup M (2019) Hepatitis C. Lancet 394(10207):1451–1466. https://doi.org/10.1016/S0140-6736(19)32320-7. (PMID: 31631857)
Benova L, Mohamoud YA, Calvert C, Abu-Raddad LJ (2014) Vertical transmission of hepatitis C virus: systematic review and meta-analysis. Clin Infect Dis 59(6):765–773
Kanninen TT, Dieterich D, Asciutti S (2015) HCV vertical transmission in pregnancy: new horizons in the era of DAAs. Hepatology 62(6):1656–1658
Dunkelberg JC, Berkley EM, Thiel KW, Leslie KK (2014) Hepatitis B and C in pregnancy: a review and recommendations for care. J Perinatol 34(12):882–891
Hellard ME, Hocking JS, Crofts N (2004) The prevalence and the risk behaviours associated with the transmission of hepatitis C virus in Australian correctional facilities. Epidemiolo Infect 132(3):409–415
Thein HH, Yi Q, Dore GJ, Krahn MD (2008) Estimation of stage-specific fibrosis progression rates in chronic hepatitis C virus infection: a meta-analysis and meta-regression. Hepatology 48(2):418–431
Thomas DL, Seeff LB (2005) Natural history of hepatitis C. Clin Liver Dis 9(3):383–398
Kallman J, O’Neil MM, Larive B, Boparai N, Calabrese L, Younossi ZM (2007) Fatigue and health-related quality of life (HRQL) in chronic hepatitis C virus infection. Dig Dis Sci 52(10):2531–2539
McLauchlan J, Lemberg MK, Hope G, Martoglio B (2002) Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J 21(15):3980–3988
Lavie M, Goffard A, Dubuisson J (2006) HCV glycoproteins: assembly of a functional E1–E2 heterodimer. In: Tan SL (ed) Hepatitis C Viruses: Genomes and Molecular Biology. Horizon Bioscience, Newyork, UK
Patel J, Patel AH, McLauchlan J (2001) The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279(1):58–68
Falkowska E, Kajumo F, Garcia E, Reinus J, Dragic T (2007) Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J Virol. https://doi.org/10.1128/JVI.00459-07
Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S (1998) Binding of hepatitis C virus to CD81. Science 282:938–941
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427.PMID:29788355
Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. https://doi.org/10.1093/nar/gkh477
Kumar A, Hossain RA, Yost SA, Bu W, Wang Y, Dearborn AD, Grakoui A, Cohen JI, Marcotrigiano J (2021) Structural insights into hepatitis C virus receptor binding and entry. Nature. https://doi.org/10.1038/s41586-021-03913-5
Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, van Gunsteren WF (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J 40(7):843–856. https://doi.org/10.1007/s00249-011-0700-9
Fischer NM, van Maaren PJ, Ditz JC, Yildirim A, van der Spoel D (2015) Properties of organic liquids when simulated with long-range lennard-jones interactions. J Chem Theory Comput 11(7):2938–2944. https://doi.org/10.1021/acs.jctc.5b00190
Tian W, Chen C, Lei X, Zhao J, Liang J (2018) CASTp 30: computed atlas of surface topography of proteins. Nucleic Acids Res. https://doi.org/10.1093/nar/gky473
Hussein HA, Borrel A, Geneix C, Petitjean M, Regad L, Camproux AC (2015) PockDrug-Server: a new web server for predicting pocket druggability on holo and apo proteins. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv462
Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S (2015) The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc. https://doi.org/10.1038/nprot.2015.043
Penkler D, Sensoy Ö, Atilgan C, Tastan BÖ (2017) Perturbation-response scanning reveals key residues for allosteric control in Hsp70. J Chem Inf Model 57(6):1359–1374. https://doi.org/10.1021/acs.jcim.6b00775
Abdizadeh H, Guven G, Atilgan AR, Atilgan C (2015) Perturbation response scanning specifies key regions in subtilisin serine protease for both function and stability. J Enzyme Inhib Med Chem 30(6):867–873. https://doi.org/10.3109/14756366.2014.979345
Hsu KC, Chen YF, Lin SR, Yang JM (2011) iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. https://doi.org/10.1186/1471-2105-12-S1-S33
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334.PMID:19499576;PMCID:PMC3041641
Liu N, Xu Z (2019) Using LeDock as a docking tool for computational drug design. IOP Conf. Ser.: Earth Environ. Sci. 218:012143
Kumari R, Kumar R, Lynn A, Open Source Drug Discovery Consortium (2014) g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962. https://doi.org/10.1021/ci500020m
https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/020482s024lbl.pdf
https://www.accessdata.fda.gov/drugsatfda_docs/label/2005/020838s022lbl.pdf
https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/200796s000lbl.pdf
https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/209445s002lbl.pdf
Hajialyani M, HoseinFarzaei M, Echeverría J, Nabavi SM, Uriarte E, Sobarzo-Sánchez E (2019) Hesperidin as a neuroprotective agent: a review of animal and clinical evidence. Molecules 24(3):648. https://doi.org/10.3390/molecules24030648
Agrawal PK, Agrawal C, Blunden G (2021) Pharmacological significance of hesperidin and hesperetin, two citrus flavonoids, as promising antiviral compounds for prophylaxis against and combating COVID-19. Nat Prod Commun. https://doi.org/10.1177/1934578X211042540
Tian S, Sun H, Pan P, Li D, Zhen X, Li Y, Hou T (2014) Assessing an ensemble docking-based virtual screening strategy for kinase targets by considering protein flexibility. J Chem Inf Model 54(10):2664–2679. https://doi.org/10.1021/ci500414b. (PMID: 25233367)
Amaro RE, Li WW (2010) Emerging methods for ensemble-based virtual screening. Curr Top Med Chem 10(1):3–13. https://doi.org/10.2174/156802610790232279
Ruan H, Yu C, Niu X, Zhang W, Liu H, Chen L, Xiong R, Sun Q, Jin C, Liu Y, Lai L (2020) Computational strategy for intrinsically disordered protein ligand design leads to the discovery of p53 transactivation domain I binding compounds that activate the p53 pathway. Chem Sci 12(8):3004–3016. https://doi.org/10.1039/d0sc04670a
Chong B, Yang Y, Zhou C, Huang Q, Liu Z (2020) Ensemble-based thermodynamics of the fuzzy binding between intrinsically disordered proteins and small-molecule ligands. J Chem Inf Model 60(10):4967–4974. https://doi.org/10.1021/acs.jcim.0c00963
Evangelista Falcon W, Ellingson SR, Smith JC, Baudry J (2019) Ensemble docking in drug discovery: how many protein configurations from molecular dynamics simulations are needed to reproduce known ligand binding? J Phys Chem B 123(25):5189–5195. https://doi.org/10.1021/acs.jpcb.8b11491
Khan M, Rauf W, Habib FE, Rahman M, Iqbal S, Shehzad A, Iqbal M (2022) Hesperidin identified from Citrus extracts potently inhibits HCV genotype 3a NS3 protease. BMC Complement Med Ther 22(1):98. https://doi.org/10.1186/s12906-022-03578-1
Mostafa TM, El-Azab GA, Badra GA, Abdelwahed AS, Elsayed AA (2021) Effect of candesartan and ramipril on liver fibrosis in patients with chronic hepatitis C viral infection: a randomized controlled prospective study. Curr Ther Res Clin Exp 95:100654. https://doi.org/10.1016/j.curtheres.2021.100654
van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des 10(3):255–262. https://doi.org/10.1007/BF00355047
Schrödinger L, DeLano W. PyMOL [Internet]. 2020. Available from: http://www.pymol.org/pymol
Yu AQ, Le J, Huang WT, Li B, Liang HX, Wang Q, Liu YT, Young CA, Zhang MY, Qin SL (2021) The effects of acarbose on non-diabetic overweight and obese patients: a meta-analysis. Adv Ther 38(2):1275–1289. https://doi.org/10.1007/s12325-020-01602-9
Hajjar I, Okafor M, Wan L, Yang Z, Nye JA, Bohsali A, Shaw LM, Levey AI, Lah JJ, Calhoun VD, Moore RH, Goldstein FC (2022) Safety and biomarker effects of candesartan in non-hypertensive adults with prodromal Alzheimer’s disease. Brain Commun 4(6):fcac270. https://doi.org/10.1093/braincomms/fcac270
Author information
Authors and Affiliations
Contributions
Protocol designed and conceptualized by DC. Manuscript preparation and data analysis done by DC, JD and SM. Project was done under the supervision of KG. The manuscript was reviewed and approved by all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chaudhuri, D., Datta, J., Majumder, S. et al. Repurposing of drug molecules from FDA database against Hepatitis C virus E2 protein using ensemble docking approach. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10646-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10646-2