Abstract
Breast cancer is recognized globally as one of the leading causes of malignant morbidity. It is a heterogeneous disease that accounts for 30 percent of all women diagnosed with cancer. To bring an anti-cancer drug from the bench to the bedside is an expensive and time-consuming process. The drug repurposing approach targets new enemies (new diseases) with old weapons (known drugs). The present study identified an FDA-approved drug targeting the γ-secretase complex involved in the Notch signaling pathway in breast cancer stem cells (BCSCs). A literature survey and in-silico study identified Venetoclax as a γ-secretase inhibitor (GSI) from 1615 FDA-approved drug compounds. In-silico docking potential of Venetoclax was better than the standard γ-secretase inhibitor RO4929097. Also, the molecular dynamics simulations of 200 ns confirmed the stability of the Venetoclax-γ-secretase complex. These findings suggest that the use of Venetoclax represents a potential γ-secretase inhibitor in breast cancer stem cells. Eventually, the in vitro and clinical evaluation will be needed to confirm the potential chemopreventive and treatment effects of Venetoclax against breast cancer and breast cancer stem cells.
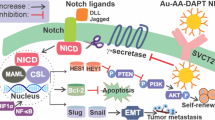
Graphical Abstract
Venetoclax appeared as the most promising drug of the 1615 FDA-approved drugs. Our in-silico findings suggest that Venetoclax may act as a γ-secretase inhibitor against the Notch signaling pathway in breast cancer stem cells.









Similar content being viewed by others
References
American Cancer Society (2021) American Cancer Society: Cancer Facts and Figures 2021. American Cancer Society, Atlanta
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin. https://doi.org/10.3322/caac.21442
Fillmore CM, Kuperwasser C (2008) Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. https://doi.org/10.1186/bcr1982
Lin Y, Zhong Y, Guan H, Zhang X, Sun Q (2012) CD44+/CD24− phenotype contributes to malignant relapse following surgical resection and chemotherapy in patients with invasive ductal carcinoma. J Exp Clin Cancer Res 31:1–9. https://doi.org/10.1186/1756-9966-31-59
Palomeras S, Ruiz-Martínez S, Puig T (2018) Targeting breast cancer stem cells to overcome treatment resistance. Molecules. https://doi.org/10.3390/molecules23092193
Shih IM, Wang TL (2007) Notch signaling, γ-secretase inhibitors, and cancer therapy. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-06-3958
Talevi A, Bellera CL (2020) Challenges and opportunities with drug repurposing: finding strategies to find alternative uses of therapeutics. Expert Opin Drug Discov. https://doi.org/10.1080/17460441.2020.1704729
Parvathaneni V, Kulkarni NS, Muth A, Gupta V (2019) Drug repurposing: a promising tool to accelerate the drug discovery process. Drug Discov Today 24:2076–2085. https://doi.org/10.1016/j.drudis.2019.06.014
Yang G, Zhou R, Guo X, Yan C, Lei J, Shi Y (2021) Structural basis of γ-secretase inhibition and modulation by small molecule drugs. Cell. https://doi.org/10.1016/j.cell.2020.11.049
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H et al (2000) The Protein Data Bank. Nucleic Acids Res. https://doi.org/10.1093/nar/28.1.235
Forli W, Halliday S, Belew R, Olson A (2012) AutoDock Version 4.2. Citeseer
Trott O, Olson AJ (2009) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. https://doi.org/10.1002/jcc.21334
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. https://doi.org/10.1002/jcc.20945
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA et al (2016) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. https://doi.org/10.1021/acs.jctc.5b00935
Lee J, Hitzenberger M, Rieger M, Kern NR, Zacharias M, Im W (2020) CHARMM-GUI supports the Amber force fields. J Chem Phys. https://doi.org/10.1063/5.0012280
Sterling T, Irwin JJ (2015) ZINC 15—ligand discovery for everyone. J Chem Inf Model. https://doi.org/10.1021/acs.jcim.5b00559J
Humphrey W, Dalke A, Schulten KVMD (1996) Visual molecular dynamics. J Mol Graph. https://doi.org/10.1016/0263-7855(96)00018-5
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Methods Mol Biol. https://doi.org/10.1007/978-1-4939-2269-7_19
Discovery Studio (2015) Dassault Systemes BIOVIA. Discovery Studio Modelling Environment, Release 4.5. Accelrys Softw Inc., San Diego
Visualizer DS (2005) v4. 0.100. 13345. Accelrys Softw. Inc., San Diego
Phillips JC, Hardy DJ, Maia JDC, Stone JE, Ribeiro JV, Bernardi RC et al (2020) Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys. https://doi.org/10.1063/5.0014475
Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O (2011) MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J Comput Chem. https://doi.org/10.1002/jcc.21787
Gowers R, Linke M, Barnoud J, Reddy T, Melo M, Seyler S et al (2016) MDAnalysis: a Python Package for the rapid analysis of molecular dynamics simulations. In: Proceedings of the 15th Python in science conference. https://doi.org/10.25080/Majora-629e541a-00e
Sanner MF, Olson AJ, Spehner JC (1995) Fast and robust computation of molecular surfaces. In: Proceedings of annual symposium on computational geometry. https://doi.org/10.1145/220279.220324
Adasme MF, Linnemann KL, Bolz SN, Kaiser F, Salentin S, Haupt VJ et al (2021) PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab294
Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZH et al (2019) End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev. https://doi.org/10.1021/acs.chemrev.9b00055
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. https://doi.org/10.1021/ar000033j
Liu H, Hou T (2016) CaFE: a tool for binding affinity prediction using end-point free energy methods. Bioinformatics. https://doi.org/10.1093/bioinformatics/btw215
Martínez L (2015) Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE. https://doi.org/10.1371/journal.pone.0119264
Tolcher AW, Messersmith WA, Mikulski SM, Papadopoulos KP, Kwak EL, Gibbon DG et al (2012) Phase I study of RO4929097, a gamma secretase inhibitor of notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J Clin Oncol. https://doi.org/10.1200/JCO.2011.36.8282
Drug Bank (2005) Amphetamine, MDMA, Metamphetamine
Acknowledgements
Part of the results presented here was developed with the help of CENAPAD-SP (Centro Nacional de Processamento de Alto Desempenho em São Paulo) grant UNICAMP/FINEP- MCT, CENAPAD-UFC (Centro Nacional de Processamento de Alto Desempenho, at Universidade Federal, do Ceará) and Digital Research Alliance of Canada (DRAC).
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
YP and IC carried out the experiment. YP wrote the manuscript with support from VT, IC, and AM. VT helped supervise the project and finally approved the version to be submitted. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest to report.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pathak, Y., Camps, I., Mishra, A. et al. Targeting notch signaling pathway in breast cancer stem cells through drug repurposing approach. Mol Divers 27, 2431–2440 (2023). https://doi.org/10.1007/s11030-022-10561-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10561-y