Abstract
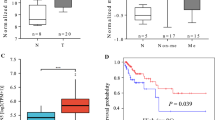
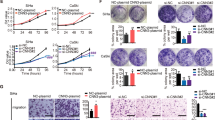
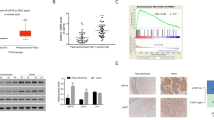
CUL4A is an ubiquitin ligase deregulated in numerous pathologies including cancer and even hijacked by viruses for facilitating their survival and propagation. However, its role in Human papilloma virus (HPV)-mediated cervical carcinogenesis remains elusive. The UALCAN and GEPIA datasets were analyzed to ascertain the transcript levels of CUL4A in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) patients. Subsequently, various biochemical assays were employed to explore the functional contribution of CUL4A in cervical carcinogenesis and to shed some light on its involvement in Cisplatin resistance in cervical cancer. Our UALCAN and GEPIA datasets analyses reveal elevated CUL4A transcript levels in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) patients that correlate with adverse clinicopathological parameters such as tumor stage and lymph node metastasis. Kaplan–Meier plot and GEPIA assessment depict poor prognosis of CESC patients having high CUL4A expression. Varied biochemical assays illustrate that CUL4A inhibition severely curtails hallmark malignant properties such as cellular proliferation, migration, and invasion of cervical cancer cells. We also show that CUL4A knockdown in HeLa cells causes increased susceptibility and better apoptotic induction toward Cisplatin, a mainstay drug used in cervical cancer treatment. More interestingly, we find reversion of Cisplatin-resistant phenotype of HeLa cells and an augmented cytotoxicity towards the platinum compound upon CUL4A downregulation. Taken together, our study underscores CUL4A as a cervical cancer oncogene and illustrates its potential as a prognosis indicator. Our investigation provides a novel avenue in improving current anti-cervical cancer therapy and overcoming the bottle-neck of Cisplatin resistance.








Similar content being viewed by others
Data availability
All data generated or analyzed in this work are included within the main article.
Abbreviations
- TCGA:
-
The Cancer Genome Atlas
- GEPIA:
-
Gene Expression Profiling Interactive Analysis
- HPV:
-
Human papillomavirus
- DNA:
-
Deoxyribonucleic acid
- CUL4A:
-
Cullin4A
- UV:
-
Ultraviolet
- HIV:
-
Human immunodeficiency virus
- HBV:
-
Hepatitis B virus
- EBV:
-
Epstein–barr virus
- SV5:
-
Simian virus 5
- HPVI2:
-
Human para influenza virus type 2
- EMT:
-
Epithelial-to-mesenchymal transition
- PARP:
-
Poly (ADP-ribose) polymerase
- pATM:
-
Phosphorylated Ataxia-telangiectasia mutated
- pChk1:
-
Phosphorylated Checkpoint kinase 1
- DSB:
-
Double-strand break
- CDK:
-
Cyclin-dependent kinase
- CDDP:
-
Cis-diamminedichloroplatinum(II)
- MTT:
-
(3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide)
- DMSO:
-
Dimethyl sulfoxide
- HCT-116:
-
Human colorectal carcinoma cells
- NER:
-
Nucleotide excision repair
- HeLa-CR:
-
HeLa-Cisplatin-resistant cells
- IAP:
-
Inhibitors of apoptosis proteins
- O.D.:
-
Optical density
- FACS:
-
Fluorescence activated cell sorting
- EDTA:
-
Ethylenediamine tetraacetic acid
- PBS:
-
Phosphate buffer saline
- γ-H2AX:
-
Phosphorylated Histone2A
References
Bray F et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Walboomers JM et al (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189(1):12–19
Schiffman MH et al (1993) Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst 85(12):958–964
McMurray HR et al (2001) Biology of human papillomaviruses. Int J Exp Pathol 82(1):15–33
Sharma P, Nag A (2014) CUL4A ubiquitin ligase: a promising drug target for cancer and other human diseases. Open Biol 4:130217
Lee J, Zhou P (2012) Pathogenic role of the CRL4 Ubiquitin ligase in Human disease. Front Oncol 2:21
Chen LC et al (1998) The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res 58(16):3677–3683
Shinomiya T et al (1999) Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer 24(4):337–344
Dohna M et al (2000) Adrenocortical carcinoma is characterized by a high frequency of chromosomal gains and high-level amplifications. Genes Chromosomes Cancer 28(2):145–152
Yasui K et al (2002) TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology 35(6):1476–1484
Michiels EM et al (2002) Genetic alterations in childhood medulloblastoma analyzed by comparative genomic hybridization. J Pediatr Hematol Oncol 24(3):205–210
Hung MS et al (2011) Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med 15(2):350–358
Ren S et al (2012) Oncogenic CUL4A determines the response to thalidomide treatment in prostate cancer. J Mol Med (Berl) 90(10):1121–1132
Yang YL et al (2014) Lung tumourigenesis in a conditional Cul4A transgenic mouse model. J Pathol 233(2):113–123
Deng J et al (2016) Cullin 4A (CUL4A), a direct target of miR-9 and miR-137, promotes gastric cancer proliferation and invasion by regulating the Hippo signaling pathway. Oncotarget 7(9):10037–10050
Birner P et al (2012) Human homologue for Caenorhabditis elegans CUL-4 protein overexpression is associated with malignant potential of epithelial ovarian tumours and poor outcome in carcinoma. J Clin Pathol 65(6):507–511
Schindl M et al (2007) Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res 27(2):949–952
Liu L et al (2009) CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell 34(4):451–460
Oh KJ et al (2004) The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin 1- and Skp2-containing E3 ligase. J Virol 78(10):5338–5346
Huh K et al (2007) Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol 81(18):9737–9747
Westrich JA et al (2018) Human papillomavirus 16 E7 stabilizes APOBEC3A protein by inhibiting cullin 2-dependent protein degradation. J Virol 92(7):e01318-17. https://doi.org/10.1128/JVI.01318-17
Mei Q et al (2020) CUL4A promotes the invasion of cervical cancer cells by regulating NF-kappaB signaling pathway. Eur Rev Med Pharmacol Sci 24(20):10403–10409
Hung MS et al (2015) Knockdown of Cul4A increases chemosensitivity to gemcitabine through upregulation of TGFBI in lung cancer cells. Oncol Rep 34(6):3187–3195
Wang Y et al (2014) CUL4A overexpression enhances lung tumor growth and sensitizes lung cancer cells to erlotinib via transcriptional regulation of EGFR. Mol Cancer 13:252
Garcia MJ et al (2013) Analysis of DNA repair-related genes in breast cancer reveals CUL4A ubiquitin ligase as a novel biomarker of trabectedin response. Mol Cancer Ther 12(4):530–541
Englinger B et al (2017) Loss of CUL4A expression is underlying cisplatin hypersensitivity in colorectal carcinoma cells with acquired trabectedin resistance. Br J Cancer 116(4):489–500
Huang G et al (2017) CUL4A overexpression as an independent adverse prognosticator in intrahepatic cholangiocarcinoma. BMC Cancer 17(1):395
Oun R, Moussa YE, Wheate NJ (2018) The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton Trans 47(19):6645–6653
Chandrashekar DS et al (2017) UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19(8):649–658
Tang Z et al (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102
Nagy A, Munkacsy G, Gyorffy B (2021) Pancancer survival analysis of cancer hallmark genes. Sci Rep 11(1):6047
John R et al (2017) Cell cycle-dependent regulation of cytoglobin by Skp2. FEBS Lett 591(21):3507–3522
Berns K et al (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428(6981):431–437
Jaiswal N et al (2015) Oncogenic human papillomavirus 16E7 modulates SUMOylation of FoxM1b. Int J Biochem Cell Biol 58:28–36
John R et al (2014) DNA damage induced activation of Cygb stabilizes p53 and mediates G1 arrest. DNA Repair (Amst) 24:107–112
Zhang L et al (2015) Butein sensitizes HeLa cells to cisplatin through the AKT and ERK/p38 MAPK pathways by targeting FoxO3a. Int J Mol Med 36(4):957–966
Duan G et al (2017) A Strategy to delay the development of cisplatin resistance by maintaining a certain amount of cisplatin-sensitive cells. Sci Rep 7(1):432
Song J, Li Y (2017) miR-25-3p reverses epithelial-mesenchymal transition via targeting Sema4C in cisplatin-resistance cervical cancer cells. Cancer Sci 108(1):23–31
Nishitani H et al (2008) CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem 283(43):29045–29052
Li B et al (2006) Cul4A targets p27 for degradation and regulates proliferation, cell cycle exit, and differentiation during erythropoiesis. Blood 107(11):4291–4299
Kotake Y, Zeng Y, Xiong Y (2009) DDB1-CUL4 and MLL1 mediate oncogene-induced p16INK4a activation. Cancer Res 69(5):1809–1814
Nag A, Bagchi S, Raychaudhuri P (2004) Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res 64(22):8152–8155
Malatesta M et al (2013) The Cul4A-DDB1 E3 ubiquitin ligase complex represses p73 transcriptional activity. Oncogene 32(39):4721–4726
Jiang L et al (2011) Cullin-4A.DNA damage-binding protein 1 E3 ligase complex targets tumor suppressor RASSF1A for degradation during mitosis. J Biol Chem 286(9):6971–8
Rosenblatt J, Gu Y, Morgan DO (1992) Human cyclin-dependent kinase 2 is activated during the S and G2 phases of the cell cycle and associates with cyclin A. Proc Natl Acad Sci U S A 89(7):2824–2828
Pines J, Hunter T (1991) Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol 115(1):1–17
Joyce NC et al (1996) Cell cycle protein expression and proliferative status in human corneal cells. Invest Ophthalmol Vis Sci 37(4):645–655
Han X et al (2016) CUL4A functions as an oncogene in ovarian cancer and is directly regulated by miR-494. Biochem Biophys Res Commun 480(4):675–681
Pan Y et al (2015) CUL4A facilitates hepatocarcinogenesis by promoting cell cycle progression and epithelial-mesenchymal transition. Sci Rep 5:17006
Wang Y et al (2014) CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res 74(2):520–531
Sui X et al (2017) CUL4A promotes proliferation and metastasis of colorectal cancer cells by regulating H3K4 trimethylation in epithelial-mesenchymal transition. Onco Targets Ther 10:735–743
Sarin N et al (2017) Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS One 12(7):e0181081
Liu Y et al (2005) The mechanism of cisplatin-induced apoptosis in HeLa cells. Chin J Clin Oncol 2(6):866–869
Lippert B (1999) Cisplatin: chemistry and biochemistry of a leading anticancer drug. Wiley, Hoboken
Martin LP, Hamilton TC, Schilder RJ (2008) Platinum resistance: the role of DNA repair pathways. Clin Cancer Res 14(5):1291–1295
Welsh C et al (2004) Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. Int J Cancer 110(3):352–361
Rosell R et al (2003) Nucleotide excision repair pathways involved in cisplatin resistance in non-small-cell lung cancer. Cancer Control 10(4):297–305
Stubbert LJ, Smith JM, McKay BC (2010) Decreased transcription-coupled nucleotide excision repair capacity is associated with increased p53- and MLH1-independent apoptosis in response to cisplatin. BMC Cancer 10:207
Rabik CA, Dolan ME (2007) Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 33(1):9–23
Stevens EV et al (2005) Expression of xeroderma pigmentosum A protein predicts improved outcome in metastatic ovarian carcinoma. Cancer 103(11):2313–2319
Weaver DA et al (2005) ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer 4(1):18
Dasari S, Tchounwou PB (2014) Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol 740:364–378
He C et al (2019) P-glycoprotein overexpression is associated with cisplatin resistance in human osteosarcoma. Anticancer Res 39(4):1711–1718
Cheng Q et al (2018) Asiatic acid (AA) sensitizes multidrug-resistant human lung adenocarcinoma A549/DDP cells to cisplatin (DDP) via downregulation of P-glycoprotein (MDR1) and its targets. Cell Physiol Biochem 47(1):279–292
Wang Y et al (2013) Involvement of CUL4A in regulation of multidrug resistance to P-gp substrate drugs in breast cancer cells. Molecules 19(1):159–176
Roy M, Mukherjee S (2014) Reversal of resistance towards cisplatin by curcumin in cervical cancer cells. Asian Pac J Cancer Prev 15(3):1403–1410
Sakaeda T et al (2002) MDR1 up-regulated by apoptotic stimuli suppresses apoptotic signaling. Pharm Res 19(9):1323–1329
Takara K et al (2006) Molecular changes to HeLa cells on continuous exposure to cisplatin or paclitaxel. Cancer Chemother Pharmacol 58(6):785–793
Okada T et al (2013) Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells. Sci Rep 3:2899
Zhu K et al (2012) Short hairpin RNA targeting Twist1 suppresses cell proliferation and improves chemosensitivity to cisplatin in HeLa human cervical cancer cells. Oncol Rep 27(4):1027–1034
Hu X et al (2019) Cul4 E3 ubiquitin ligase regulates ovarian cancer drug resistance by targeting the antiapoptotic protein BIRC3. Cell Death Dis 10(2):104
Acknowledgements
We thank CIF, UDSC, for providing FACS facility
Funding
This work was supported by Faculty Research Programme Grant from Institution of Eminence, University of Delhi (lOE/FRP/LS/2020/27), DU-DST (PURSE Grant: RC/2014/7114), UGC-SAP Grant (F.3.3./2016 DRS II UGC-SAP II), CSIR EMR Grant (37(1682)/17/EMR-II), and DBT Grant (BT/PR15422/MED/30/1705/2016) to AN. Financial assistance from SERB/JC Bose program Ref# SR/S2/JCB-08/2010 to DPS is duly acknowledged. Fellowships were provided by UGC to YA, CSIR to HB, and DBT to NS.
Author information
Authors and Affiliations
Contributions
YA contributed to conceptualization, methodology, data curation, validation, investigation, formal analysis, writing: original draft, and visualization. HB contributed to methodology, data curation, formal analysis, writing: review and editing, and visualization. NS contributed to data curation during revision, data analysis, writing: review. DPS contributed to writing: review and editing and funding acquisition. AN conceptualization, methodology, resources, writing: review and editing, supervision, project administration, and funding acquisition. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Atri, Y., Bharti, H., Sahani, N. et al. CUL4A silencing attenuates cervical carcinogenesis and improves Cisplatin sensitivity. Mol Cell Biochem 479, 1041–1058 (2024). https://doi.org/10.1007/s11010-023-04776-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-023-04776-2