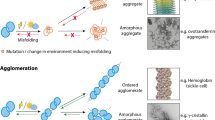
Abstract
Proteins are one of the dynamic macromolecules that play a significant role in many physiologically important processes to sustain life on the earth. Proteins need to be properly folded into their active conformation to perform their function. Alteration in the protein folding process may lead to the formation of misfolded conformers. Accumulation of these misfolded conformers can result in the formation of protein aggregates which are attributed to many human pathological conditions including neurodegeneration, cataract, neuromuscular disorders, and diabetes. Living cells naturally have heterogeneous crowding environments with different concentrations of various biomolecules. Macromolecular crowding condition has been found to alter the protein conformation. Here in this review, we tried to show the relation between macromolecular crowding, protein aggregation, and its consequences.





Similar content being viewed by others
Availability of Data and Materials
Not applicable.
Code Availability
Not applicable.
References
Van Den Berg J, Boersma AJ, Poolman B (2017) Microorganisms maintain crowding homeostasis. Nat Rev Microbiol 15:309–318
Miklos AC, Sarkar M, Wang Y, Pielak GJ (2011) Protein crowding tunes protein stability. J Am Chem Soc 133:7116–7120
Zimmerman SB, Trach SO (1991) Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol 222:599–620
Szathmáry E, Smith JM (1995) The major evolutionary transitions. Nature 374:227–232
Ellis RJ (2001) Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr Opin Struct Biol 11:114–119
Harding J (1991) Biochemistry epidemiology and pharmacology. Cataract 195–217
Minton AP, Wilf J (1981) Effect of macromlecular crowding upon the structure and function of an enzyme: glyceraldehyde-3-phosphate dehydrogenase. Biochemistry 20:4821–4826
Christiansen A, Wang Q, Samiotakis A et al (2010) Factors defining effects of macromolecular crowding on protein stability: an in vitro/in silico case study using cytochrome c. Biochemistry 49:6519–6530
Mittal S, Singh LR (2014) Macromolecular crowding induces holo α-lactalbumin aggregation by converting to its apo form. PLoS ONE 9:e114029
Du F, Zhou Z, Mo ZY et al (2006) Mixed macromolecular crowding accelerates the refolding of rabbit muscle creatine kinase: implications for protein folding in physiological environments. J Mol Biol 364:469–482
Kozer N, Kuttner YY, Haran G, Schreiber G (2007) Protein-protein association in polymer solutions: from dilute to semidilute to concentrated. Biophys J 92:2139–2149
Wenner JR, Bloomfield VA (1999) Crowding effects on EcoRV kinetics and binding. Biophys J 77:3234–3241
Rivas G, Fernandez JA, Minton AP (1999) Direct observation of the self-association of dilute proteins in the presence of inert macromolecules at high concentration via tracer sedimentation equilibrium: theory, experiment, and biological significance. Biochemistry 38:9379–9388
Verma PK, Rakshit S, Mitra RK, Pal SK (2011) Role of hydration on the functionality of a proteolytic enzyme α-chymotrypsin under crowded environment. Biochimie 93:1424–1433
Mittal S, Chowhan RK, Singh LR (2015) Macromolecular crowding: Macromolecules friend or foe. Biochim Biophys Acta (BBA)-Gen Subj 1850:1822–1831
Mittal S, Singh LR (2013) Denatured state structural property determines protein stabilization by macromolecular crowding: a thermodynamic and structural approach. PLoS ONE 8:e78936
Stagg L, Zhang S-Q, Cheung MS, Wittung-Stafshede P (2007) Molecular crowding enhances native structure and stability of α/β protein flavodoxin. Proc Natl Acad Sci 104:18976–18981
Roque A, Ponte I, Suau P (2007) Macromolecular crowding induces a molten globule state in the C-terminal domain of histone H1. Biophys J 93:2170–2177
Perham M, Stagg L, Wittung-Stafshede P (2007) Macromolecular crowding increases structural content of folded proteins. FEBS Lett 581:5065–5069
Cheung MS, Klimov D, Thirumalai D (2005) Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc Natl Acad Sci U S A 102:4753–4758
Muramatsu N, Minton AP (1988) Tracer diffusion of globular proteins in concentrated protein solutions. Proc Natl Acad Sci 85:2984–2988
Han J, Herzfeld J (1993) Macromolecular diffusion in crowded solutions. Biophys J 65:1155–1161
Dhar A, Samiotakis A, Ebbinghaus S et al (2010) Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc Natl Acad Sci 107:17586–17591
Derham BK, Harding JJ (2006) The effect of the presence of globular proteins and elongated polymers on enzyme activity. Biochim Biophys Acta (BBA)-Proteins Proteomics 1764:1000–1006
Jiang M, Guo Z (2007) Effects of macromolecular crowding on the intrinsic catalytic efficiency and structure of enterobactin-specific isochorismate synthase. J Am Chem Soc 129:730–731
Norris MGS, Malys N (2011) What is the true enzyme kinetics in the biological system? An investigation of macromolecular crowding effect upon enzyme kinetics of glucose-6-phosphate dehydrogenase. Biochem Biophys Res Commun 405:388–392
Mittal S, Singh LR (2014) Macromolecular crowding decelerates aggregation of a β-rich protein, bovine carbonic anhydrase: a case study. J Biochem 156:273–282
Ma B, Xie J, Wei L, Li W (2013) Macromolecular crowding modulates the kinetics and morphology of amyloid self-assembly by β-lactoglobulin. Int J Biol Macromol 53:82–87
Siddiqui GA, Naeem A (2018) Aggregation of globular protein as a consequences of macromolecular crowding: A time and concentration dependent study. Int J Biol Macromol 108:360–366
Siddiqui GA, Naeem A (2020) The contrasting effect of macromolecular crowding and confinement on fibril formation of globular protein: Underlying cause of proteopathies. J Mol Liq 114602
van den Berg B, Ellis RJ, Dobson CM (1999) Effects of macromolecular crowding on protein folding and aggregation. EMBO J 18:6927–6933
van den Berg B, Wain R, Dobson CM, Ellis RJ (2000) Macromolecular crowding perturbs protein refolding kinetics: implications for folding inside the cell. EMBO J 19:3870–3875
Fan Y-Q, Liu H-J, Li C et al (2012) Effects of macromolecular crowding on refolding of recombinant human brain-type creatine kinase. Int J Biol Macromol 51:113–118
Malik A, Kundu J, Mukherjee SK, Chowdhury PK (2012) Myoglobin unfolding in crowding and confinement. J Phys Chem B 116:12895–12904
Huang L, Jin R, Li J et al (2010) Macromolecular crowding converts the human recombinant PrPC to the soluble neurotoxic β-oligomers. FASEB J 24:3536–3543
Harada R, Tochio N, Kigawa T et al (2013) Reduced native state stability in crowded cellular environment due to protein–protein interactions. J Am Chem Soc 135:3696–3701
Sarkar M, Smith AE, Pielak GJ (2013) Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci 110:19342–19347
Chi EY, Krishnan S, Randolph TW, Carpenter JF (2003) Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm Res 20:1325–1336
Castillo V, Graña-Montes R, Sabate R, Ventura S (2011) Prediction of the aggregation propensity of proteins from the primary sequence: aggregation properties of proteomes. Biotechnol J 6:674–685
Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Med 10:S10–S17
Kholova I, Niessen HWM (2005) Amyloid in the cardiovascular system: a review. J Clin Pathol 58:125–133
Williams RA, Mamotte CDS, Burnett JR (2008) Phenylketonuria: an inborn error of phenylalanine metabolism. Clin Biochem Rev 29:31
Xu J, Reumers J, Couceiro JR et al (2011) Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 7:285–295
Chowhan RK, Warepam M, Dar TA, Singh LR (2013) Recent trends in treating neuronal proteinopathies. J Proteins Proteomics 4
Chowhan RK, Mittal S, Dar TA et al (2014) Ignored avenues in alpha-synuclein associated proteopathy. CNS Neurol Disord Targets (Formerly Curr Drug Targets-CNS Neurol Disord) 13:1246–1257
Ahmed A, Shamsi A, Bano B (2017) Characterizing harmful advanced glycation end-products (AGEs) and ribosylated aggregates of yellow mustard seed phytocystatin: Effects of different monosaccharides. Spectrochim Acta Part A Mol Biomol Spectrosc 171:183–192
Galán A, Sot B, Llorca O et al (2001) Excluded volume effects on the refolding and assembly of an oligomeric protein: GroEL, a case study. J Biol Chem 276:957–964
Zhou BR, Liang Y, Du F et al (2004) Mixed macromolecular crowding accelerates the oxidative refolding of reduced, denatured lysozyme: Implications for protein folding in intracellular environments. J Biol Chem 279:55109–55116
Munishkina LA, Ahmad A, Fink AL, Uversky VN (2008) Guiding protein aggregation with macromolecular crowding. Biochemistry 47:8993–9006
Ma Q, Fan J-B, Zhou Z et al (2012) The contrasting effect of macromolecular crowding on amyloid fibril formation. PLoS ONE 7:e36288
Hatters DM, Minton AP, Howlett GJ (2002) Macromolecular crowding accelerates amyloid formation by human apolipoprotein C-II. J Biol Chem 277:7824–7830
Ghahghaei A, Divsalar A, Faridi N (2010) The effects of molecular crowding on the amyloid fibril formation of α-lactalbumin and the chaperone action of α-casein. Protein J 29:257–264
Zhou Z, Fan J-B, Zhu H-L et al (2009) Crowded cell-like environment accelerates the nucleation step of amyloidogenic protein misfolding. J Biol Chem 284:30148–30158
Yamin G, Munishkina LA, Karymov MA et al (2005) Forcing nonamyloidogenic β-synuclein to fibrillate. Biochemistry 44:9096–9107
Munishkina LA, Cooper EM, Uversky VN, Fink AL (2004) The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J Mol Recognit 17:456–464
Uversky VN, Cooper EM, Bower KS et al (2002) Accelerated α-synuclein fibrillation in crowded milieu. Febs Lett 515:99–103
Homchaudhuri L, Sarma N, Swaminathan R (2006) Effect of crowding by dextrans and Ficolls on the rate of alkaline phosphatase–catalyzed hydrolysis: A size-dependent investigation. Biopolym Orig Res Biomol 83:477–486
Gellerich FN, Laterveer FD, Korzeniewski B et al (1998) Dextran strongly increases the Michaelis constants of oxidative phosphorylation and of mitochondrial creatine kinase in heart mitochondria. Eur J Biochem 254:172–180
Pastor I, Vilaseca E, Madurga S et al (2011) Effect of crowding by dextrans on the hydrolysis of N-succinyl-l-phenyl-ala-p-nitroanilide catalyzed by α-chymotrypsin. J Phys Chem B 115:1115–1121
Ren G, Lin Z, Tsou C, Wang C (2003) Effects of macromolecular crowding on the unfolding and the refolding of D-glyceraldehyde-3-phosophospate dehydrogenase. J Protein Chem 22:431–439
Levy Y, Onuchic JN (2004) Water and proteins: A love–hate relationship. Proc Natl Acad Sci 101:3325–3326
Rupley JA, Careri G (1991) Protein hydration and function. Adv Protein Chem 41:37–172
Frölich A, Gabel F, Jasnin M et al (2009) From shell to cell: neutron scattering studies of biological water dynamics and coupling to activity. Faraday Discuss 141:117–130
Mansell JL, Clegg JS (1983) Cellular and molecular consequences of reduced cell water content. Cryobiology 20:591–612
Harada R, Sugita Y, Feig M (2012) Protein crowding affects hydration structure and dynamics. J Am Chem Soc 134:4842–4849
Yaku H, Murashima T, Tateishi-Karimata H et al (2013) Study on effects of molecular crowding on G-quadruplex-ligand binding and ligand-mediated telomerase inhibition. Methods 64:19–27
Shah H, Rawat K, Ashar H et al (2019) Dual role for fungal-specific outer kinetochore proteins during cell cycle and development in Magnaporthe oryzae. J Cell Sci 132:jcs224147
Radwan M, Wood RJ, Sui X, Hatters DM (2017) When proteostasis goes bad: protein aggregation in the cell. IUBMB Life 69:49–54
Stefani M, Dobson CM (2003) Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med 81:678–699
Gregersen N, Bolund L, Bross P (2005) Protein misfolding, aggregation, and degradation in disease. Mol Biotechnol 31:141–150
Ilyinsky NS, Nesterov SV, Shestoperova EI et al (2021) On the role of normal aging processes in the onset and pathogenesis of diseases associated with the abnormal accumulation of protein aggregates. Biochem 86:275–289
Sayre LM, Perry G, Smith MA (2008) Oxidative stress and neurotoxicity. Chem Res Toxicol 21:172–188
Sweeney P, Park H, Baumann M et al (2017) Protein misfolding in neurodegenerative diseases: implications and strategies. Transl Neurodegener 6:1–13
Gregersen N, Bross P (2010) Protein misfolding and cellular stress: an overview. Protein Misfolding Cell Stress Dis Aging 3–23
Furkan M, Siddiqi MK, Zakariya SM et al (2019) An In Vitro elucidation of the antiaggregatory potential of Diosminover thermally induced unfolding of hen egg white lysozyme; A preventive quest for lysozyme amyloidosis. Int J Biol Macromol 129:1015–1023
Arfin S, Siddiqui GA, Naeem A, Moin S (2018) Inhibition of advanced glycation end products by isoferulic acid and its free radical scavenging capacity: An in vitro and molecular docking study. Int J Biol Macromol 118:1479–1487
Fazili NA, Siddiqui GA, Bhat SA et al (2015) Rifampicin induced aggregation of ovalbumin: Malicious behaviour of antibiotics. Protein Pept Lett 22
Knaupp AS, Bottomley SP (2009) Serpin polymerization and its role in disease—The molecular basis of α1-antitrypsin deficiency. IUBMB Life 61:1–5
Amani S, Naeem A (2013) Understanding protein folding from globular to amyloid state: aggregation: darker side of protein. Process Biochem 48:1651–1664
Meersman F, Dobson CM (2006) Probing the pressure–temperature stability of amyloid fibrils provides new insights into their molecular properties. Biochim Biophys Acta (BBA)-Proteins Proteomics 1764:452–460
Brundin P, Melki R, Kopito R (2010) Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol cell Biol 11:301–307
Moreno-Gonzalez I, Soto C (2011) Misfolded protein aggregates: mechanisms, structures and potential for disease transmission. In Seminars in cell & developmental biology. Elsevier, pp 482–487
Jucker M, Walker LC (2013) Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501:45–51
Davis AA, Leyns CEG, Holtzman DM (2018) Intercellular spread of protein aggregates in neurodegenerative disease. Annu Rev Cell Dev Biol 34:545–568
Blancas-Mejía LM, Ramirez-Alvarado M (2013) Systemic amyloidoses. Annu Rev Biochem 82:745–774
Lee S-J, Desplats P, Sigurdson C et al (2010) Cell-to-cell transmission of non-prion protein aggregates. Nat Rev Neurol 6:702–706
Bucciantini M, Giannoni E, Chiti F et al (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416:507–511
Jiang T, Yu J-T, Tian Y, Tan L (2013) Epidemiology and etiology of Alzheimer’s disease: from genetic to non-genetic factors. Curr Alzheimer Res 10:852–867
Monsellier E, Ramazzotti M, Taddei N, Chiti F (2008) Aggregation propensity of the human proteome. PLoS Comput Biol 4:e1000199
Hensley K, Carney JM, Mattson MP et al (1994) A model for beta-amyloid aggregation and neurotoxicity based on free radical generation by the peptide: relevance to Alzheimer disease. Proc Natl Acad Sci 91:3270–3274
Siddiqui GA, Siddiqi MK, Khan RH, Naeem A (2018) Probing the binding of phenolic aldehyde vanillin with bovine serum albumin: Evidence from spectroscopic and docking approach. Spectrochim Acta - Part A Mol Biomol Spectrosc 203:40–47
Dawn A, Deep S (2020) Thinking beyond tradition: Polyphenols as effective refolding modulators. Int J Biol Macromol 148:969–978
Acknowledgements
The authors would like to acknowledge the facilities obtained at Aligarh Muslim University, Aligarh.
Author information
Authors and Affiliations
Contributions
Review article written by Gufran Ahmed Siddiqui and checked by Dr Aabgeena Naeem.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declared that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Siddiqui, G.A., Naeem, A. Connecting the Dots: Macromolecular Crowding and Protein Aggregation. J Fluoresc 33, 1–11 (2023). https://doi.org/10.1007/s10895-022-03082-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-022-03082-2