Abstract
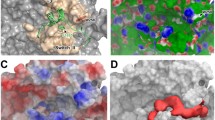
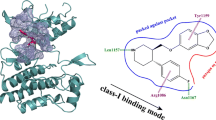
We used NMR to show that the antipsychotic phenothiazine drugs promazine and promethazine bind to GDP-KRAS. Promazine also binds to oncogenic GDP-KRAS(G12D), and to wild type GppNHp-KRAS. A panel of additional phenothiazines bind to GDP-KRAS but with lower affinity than promazine or promethazine. Binding is most dependent on substitutions at C-2 of the tricyclic phenothiazine ring. Promazine was used to generate an NMR-driven HADDOCK model of the drug/GDP-KRAS complex. The structural model shows the tricyclic phenothiazine ring of promazine associates with the hydrophobic pocket p1 that is bordered by the central β sheet and Switch II in KRAS. Binding appears to stabilize helix 2 in a conformation that is similar to that seen in KRAS bound to other small molecules. Association of phenothiazines with KRAS may affect normal KRAS signaling that could contribute to multiple biological activities of these antipsychotic drugs. Moreover, the phenothiazine ring represents a new core scaffold on which to design modulators of KRAS activity.







Similar content being viewed by others
Availability of data and material
The atomic coordinates of the final ensemble of 10 structure models have been deposited in the Protein Data Bank (PDB ID: 7LGI) and in the Biological Magnetic Resonance Bank (BMRB entry ID: 30845). Authors will release the atomic coordinates and experimental data upon article publication.
References
Aftab DT, Ballas LM, Loomis CR, Hait WN (1991) Structure-activity relationships of phenothiazines and related drugs for inhibition of protein kinase C. Mol Pharmacol 40:798–805
Araki M et al (2011) Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers. J Biol Chem 286:39644–39653
Attwood D, Florence AT, Gillan JM (1974) Micellar properties of drugs: properties of micellar aggregates of phenothiazines and their aqueous solutions. J Pharm Sci 63:988–993
Azuine MA et al (2004) Cancer chemopreventive effect of phenothiazines and related tri-heterocyclic analogues in the 12-O-tetradecanoylphorbol-13-acetate promoted Epstein-Barr virus early antigen activation and the mouse skin two-stage carcinogenesis models. Pharmacol Res 49:161–169
Baciu-Atudosie L et al (2012) Synthesis and biological evaluation of new phenothiazine derivatives bearing a pyrazole unit as protein farnesyltransferase inhibitors. Bioorg Med Chem Lett 22:6896–6902
Baral PK et al (2014) Structural basis of prion inhibition by phenothiazine compounds. Structure 22:291–303
Breeze AL (2000) Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog NMR Spectrosc 36:323–372
Buhrman G et al (2011) Analysis of binding site hot spots on the surface of Ras GTPase. J Mol Biol 413:773–789
Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ (2014) Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 13:828–851
Delaglio F et al (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Eisenberg S, Giehl K, Henis YI, Ehrlich M (2008) Differential interference of chlorpromazine with the membrane interactions of oncogenic K-Ras and its effects on cell growth. J Biol Chem 283:27279–27288
Fang Z et al (2018) Inhibition of K-RAS4B by a unique mechanism of action: stabilizing membrane-dependent occlusion of the effector-binding site. Cell Chem Biol 25:1327–1336
Florence AT, Parfitt RT (1971) Micelle formation by some phenothiazine derivatives. II. Nuclear magnetic resonance studies in deuterium oxide. J Phys Chem 75:3554–3560
Grant BJ et al (2011) Novel allosteric sites on Ras for lead generation. PLoS ONE 6:
Ito Y et al (1997) Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry 36:9109–9119
Jansen JM et al (2017) Inhibition of prenylated KRAS in a lipid environment. PLoS ONE 12:e0174706
Jaszczyszyn A et al (2012) Chemical structure of phenothiazines and their biological activity. Pharmacol Rep 64:16–23
Johnson BA, Blevins RA (1994) NMR view: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4:603–614
Johnson CW et al (2017) The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects. J Biol Chem 292:12981–12993
Mao Y, Yao H, Wang H, Cheng P, Long D (2016) Microsecond timescale dynamics of GDP-bound Ras underlies the formation of novel inhibitor-binding pockets. Angew Chem Int Ed Engl 55:15629–15632
Marshall CB et al (2020) NMR in intregrated biophysical drug discovery for RAS: past, present and future. J Biomol NMR 74:531–554
Massom L, Lee H, Jarrett HW (1990) Trifluoperazine binding to porcine brain calmodulin and skeletal muscle troponin C. Biochemistry 29:671–681
Maurer T et al (2012) Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA 109:5299–5304
Meyer B et al (2004) Saturation transfer difference NMR spectroscopy for identifying ligand epitopes and binding specificities. Ernst Schering Res Found Workshop, pp 149–67
Motohashi N, Kawase M, Satoh K, Sakagami H (2006) Cytotoxic potential of phenothiazines. Curr Drug Targets 7:1055–1066
O’Connor C, Kovrigin EL (2012) Assignments of backbone (1)H, (1)(3)C and (1)(5)N resonances in H-Ras (1-166) complexed with GppNHp at physiological pH. Biomol NMR Assign 6:91–93
Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503:548–551
Palfy G, Vlda I, Perczel A (2020) 1H,15 N backbone assignment and comparative analysis of the wild type and G12C, G12D, G12V mutants of K-Ras bound to GDP ar physiological pH. Biomol NMR Assign 14:1–7
Parker JA, Volmar AY, Pavlopoulos S, Mattos C (2018) K-Ras populates conformational states differently from its isoform H-Ras and oncogenic mutant K-RasG12D. Structure 26(6):810–820
Pluta K, Morak-Mlodawska B, Jelen M (2011) Recent progress in biological activities of synthesized phenothiazines. Eur J Med Chem 46:3179–3189
Prior IA, Lewis PD, Mattos C (2012) A comprehensive suvey of Ras mutations in cancer. Cancer Res 72:2457–2467
Ramu A, Ramu N (1992) Reversal of multidrug resistance by phenothiazines and structurally related compounds. Cancer Chemother Pharmacol 30:165–173
Rodrigues JP, Karaca E, Bonvin AM (2015) Information-driven structural modelling of protein-protein interactions. Methods Mol Biol 1215:399–424
Shima F et al (2010) Structural basis for conformational dynamics of GTP-bound Ras protein. J Biol Chem 285:22696–22705
Shima F et al (2013) In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci USA 110:8182–8187
Spengler G et al (2014) Multidrug resistance reversing activity of newly developed phenothiazines on P-glycoprotein (ABCB1)-related resistance of mouse T-lymphoma cells. Anticancer Res 34:1737–1741
Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H (2014) Small-molecule modulation of Ras signaling. Nat Chem Biol 10:613–622
Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A (2001) Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA 98:4944–4949
Sun Q et al (2012) Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl 51:6140–6143
Vandonselaar M, Hickie RA, Quail JW, Delbaere LTJ (1994) Trifluoperazine-induced conformational changes in Ca2 + -calmodulin. Nat Struct Biol 1:795–801
Varga B et al (2017) Possible biological and clinical applications of phenothiazines. Anticancer Res 37:5983–5993
Vo U, Embrey KJ, Breeze AL, Golovanov AP (2013) (1)H, (1)(3)C and (1)(5)N resonance assignment for the human K-Ras at physiological pH. Biomol NMR Assign 7:215–219
Wagstaff JL, Taylor SL, Howard MJ (2013) Recent developments and applications of saturation transfer difference nuclear magnetic resonance (STD NMR) spectroscopy. Mol BioSyst 9:571–577
Welsch ME et al (2017) Multivalent small-molecule PAN-RAS inhibitors. Cell 168:878–889
Wishart DS et al (1995) 1H, 13C and 15 N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140
Wu CH et al (2016) Pharmacological exploitation of the phenothiazine antipsychotics to develop novel antitumor agents-A drug repurposing strategy. Sci Rep 6:27540
Xu S et al (2017) Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras. Acta Crystallogr D Struct Biol 73:970–984
Funding
This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT Grant No DP150093).
Author information
Authors and Affiliations
Contributions
X.W. performed and analyzed the majority of all NMR experiments, helped prepare figures, wrote the initial draft and proof-read the paper. A.A.G. helped design the experiments and proof-read the manuscript. J.A.P. supervised all experiments, performed some of the NMR experiments, wrote the final drafts, prepared the final figures, and proof read the final draft. All authors have given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Consent to publication
All authors have read the manuscript and evaluated the data, and agree to publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, X., Gorfe, A.A. & Putkey, J.A. Antipsychotic phenothiazine drugs bind to KRAS in vitro. J Biomol NMR 75, 233–244 (2021). https://doi.org/10.1007/s10858-021-00371-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-021-00371-z