Abstract
Purpose
The preimplantation genetic testing for aneuploidy (PGT-A) platform is not currently available for small copy-number variants (CNVs), especially those < 1 Mb. Through strategies used in PGT for monogenic disease (PGT-M), this study intended to perform PGT for families with small pathogenic CNVs.
Methods
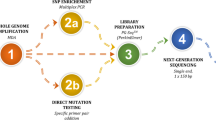
Couples who carried small pathogenic CNVs and underwent PGT at the Reproductive and Genetic Hospital of CITIC-Xiangya (Hunan, China) between November 2019 and April 2023 were included in this study. Haplotype analysis was performed through two platforms (targeted sequencing and whole-genome arrays) to identify the unaffected embryos, which were subjected to transplantation. Prenatal diagnosis using amniotic fluid was performed during 18–20 weeks of pregnancy.
Results
PGT was successfully performed for 20 small CNVs (15 microdeletions and 5 microduplications) in 20 families. These CNVs distributed on chromosomes 1, 2, 6, 7, 13, 15, 16, and X with sizes ranging from 57 to 2120 kb. Three haplotyping-based PGT-M strategies were applied. A total of 89 embryos were identified in 25 PGT cycles for the 20 families. The diagnostic yield was 98.9% (88/89). Nineteen transfers were performed for 17 women, resulting in a 78.9% (15/19) clinical pregnancy rate after each transplantation. Of the nine women who had healthy babies, eight accepted prenatal diagnosis and the results showed no related pathogenic CNVs.
Conclusion
Our results show that the extended haplotyping-based PGT-M strategy application for small pathogenic CNVs compensated for the insufficient resolution of PGT-A. These three PGT-M strategies could be applied to couples with small pathogenic CNVs.



Similar content being viewed by others
Data availability
The targeted sequencing or SNP array data in this study have not been deposited in public repository because of privacy and ethical restrictions but are available from the corresponding author on request.
References
Watson CT, Marques-Bonet T, Sharp AJ, Mefford HC. The genetics of microdeletion and microduplication syndromes: an update. Annu Rev Genomics Hum Genet. 2014;15:215–44. https://doi.org/10.1146/annurev-genom-091212-153408.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22(2):245–57. https://doi.org/10.1038/s41436-019-0686-8.
Ledbetter DH, Riccardi VM, Airhart SD, Strobel RJ, Keenan BS, Crawford JD. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N Engl J Med. 1981;304(6):325–9. https://doi.org/10.1056/NEJM198102053040604.
Knoll JH, Nicholls RD, Magenis RE, Graham JM Jr, Lalande M, Latt SA. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet. 1989;32(2):285–90. https://doi.org/10.1002/ajmg.1320320235.
Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14(10):417–22. https://doi.org/10.1016/s0168-9525(98)01555-8.
Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1(6):e49. https://doi.org/10.1371/journal.pgen.0010049.
Chau MHK, Cao Y, Kwok YKY, Chan S, Chan YM, Wang H, et al. Characteristics and mode of inheritance of pathogenic copy number variants in prenatal diagnosis. Am J Obstet Gynecol. 2019;221(5):493e1. https://doi.org/10.1016/j.ajog.2019.06.007.
De Rycke M, Berckmoes V. Preimplantation genetic testing for monogenic disorders. Genes (Basel). 2020;11(8) https://doi.org/10.3390/genes11080871.
ESHRE PGT Consortium Steering Committee, Carvalho F, Coonen E, Goossens V, Kokkali G, Rubio C, et al. ESHRE PGT Consortium good practice recommendations for the organisation of PGT. Hum Reprod Open. 2020;2020(3):hoaa021. https://doi.org/10.1093/hropen/hoaa021.
Geraedts J, Sermon K. Preimplantation genetic screening 2.0: the theory. Mol Hum Reprod. 2016;22(8):839–44. https://doi.org/10.1093/molehr/gaw033.
Xie P, Liu P, Zhang S, Cheng D, Chen D, Tan YQ, et al. Segmental aneuploidies with 1 Mb resolution in human preimplantation blastocysts. Genet Med. 2022;24(11):2285–95. https://doi.org/10.1016/j.gim.2022.08.008.
ESHRE PGT-M Working Group, Carvalho F, Moutou C, Dimitriadou E, Dreesen J, Gimenez C, et al. ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders. Hum Reprod Open. 2020;2020(3):hoaa018. https://doi.org/10.1093/hropen/hoaa018.
Sabria-Back J, Monteagudo-Sanchez A, Sanchez-Delgado M, Ferguson-Smith AC, Gomez O, Pertierra Cartada A, et al. Preimplantation genetic testing for a chr14q32 microdeletion in a family with Kagami-Ogata syndrome and Temple syndrome. J Med Genet. 2022;59(3):253–61. https://doi.org/10.1136/jmedgenet-2020-107433.
Rotshenker-Olshinka K, Srebnik Moshe N, Weiss O, Shaviv S, Freireich O, Segel R, et al. Preimplantation genetic testing (PGT) for copy number variants of uncertain significance (CNV- VUS) in the genomic era: to do or not to do? J Assist Reprod Genet. 2021;38(3):719–25. https://doi.org/10.1007/s10815-020-02055-3.
Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367(14):1321–31. https://doi.org/10.1056/NEJMoa1200395.
Lee I, Alur-Gupta S, Gallop R, Dokras A. Utilization of preimplantation genetic testing for monogenic disorders. Fertil Steril. 2020;114(4):854–60. https://doi.org/10.1016/j.fertnstert.2020.05.045.
Zhang S, Lei C, Wu J, Xiao M, Zhou J, Zhu S, et al. A comprehensive and universal approach for embryo testing in patients with different genetic disorders. Clin Transl Med. 2021;11(7):e490. https://doi.org/10.1002/ctm2.490.
Hu X, He WB, Zhang SP, Luo KL, Gong F, Dai J, et al. Next-generation sequence-based preimplantation genetic testing for monogenic disease resulting from maternal mosaicism. Mol Genet Genomic Med. 2021;9(5):e1662. https://doi.org/10.1002/mgg3.1662.
Zhang S, Lei C, Wu J, Zhou J, Sun H, Fu J, et al. The establishment and application of preimplantation genetic haplotyping in embryo diagnosis for reciprocal and robertsonian translocation carriers. BMC Med Genomics. 2017;10(1):60.https://doi.org/10.1186/s12920-017-0294-x
Xie P, Hu X, Kong L, Mao Y, Cheng D, Kang K, et al. A novel multifunctional haplotyping-based preimplantation genetic testing for different genetic conditions. Hum Reprod. 2022;37(11):2546–59. https://doi.org/10.1093/humrep/deac190.
Zhou S, Xie P, Zhang S, Hu L, Luo K, Gong F, et al. Complex mosaic blastocysts after preimplantation genetic testing: prevalence and outcomes after re-biopsy and re-vitrification. Reprod Biomed Online. 2021;43(2):215–22. https://doi.org/10.1016/j.rbmo.2021.04.006.
Yuan S, Guo L, Cheng D, Li X, Hu H, Hu L, et al. The de novo aberration rate of prenatal karyotype was comparable between 1496 fetuses conceived via IVF/ICSI and 1396 fetuses from natural conception. J Assist Reprod Genet. 2022;39(7):1683–9. https://doi.org/10.1007/s10815-022-02500-5.
Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2013;15(6):478–81. https://doi.org/10.1038/gim.2012.164.
Kirov G. CNVs in neuropsychiatric disorders. Hum Mol Genet. 2015;24(R1):R45–9. https://doi.org/10.1093/hmg/ddv253.
Liu Z, Yin N, Gong L, Tan Z, Yin B, Yang Y, et al. Microduplication of 7q36.3 encompassing the SHH long-range regulator (ZRS) in a patient with triphalangeal thumb-polysyndactyly syndrome and congenital heart disease. Mol Med Rep. 2017;15(2):793–7. https://doi.org/10.3892/mmr.2016.6092.
Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84(2):148–61. https://doi.org/10.1016/j.ajhg.2008.12.014.
Capalbo A, Rienzi L, Ubaldi FM. Diagnosis and clinical management of duplications and deletions. Fertil Steril. 2017;107(1):12–8. https://doi.org/10.1016/j.fertnstert.2016.11.002.
Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. https://doi.org/10.1146/annurev-genom-090413-025352.
Dittwald P, Gambin T, Szafranski P, Li J, Amato S, Divon MY, et al. NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Res. 2013;23(9):1395–409. https://doi.org/10.1101/gr.152454.112.
Tu J, Zhou Y, Tao Y, Lu N, Yang Y, Lu Z. Sensitivity to copy number variation analysis in single cell genomics. Gene. 2022;808:145995. https://doi.org/10.1016/j.gene.2021.145995.
Hughes T, Bracewell-Milnes T, Saso S, Jones BP, Almeida PA, Maclaren K, et al. A review on the motivations, decision-making factors, attitudes and experiences of couples using pre-implantation genetic testing for inherited conditions. Hum Reprod Update. 2021;27(5):944–66. https://doi.org/10.1093/humupd/dmab013.
Zschocke J, Byers PH, Wilkie AOM. Mendelian inheritance revisited: dominance and recessiveness in medical genetics. Nat Rev Genet. 2023.https://doi.org/10.1038/s41576-023-00574-0.
Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013;132(10):1077–130. https://doi.org/10.1007/s00439-013-1331-2.
Di-Battista A, Meloni VA, da Silva MD, Moyses-Oliveira M, Melaragno MI. Unusual X-chromosome inactivation pattern in patients with Xp11.23-p11.22 duplication: Report and review. Am J Med Genet A. 2016;170(12):3271–5. https://doi.org/10.1002/ajmg.a.37888.
Payne JL. Psychopharmacology in pregnancy and breastfeeding. Med Clin North Am. 2019;103(4):629–50. https://doi.org/10.1016/j.mcna.2019.02.009.
Acknowledgements
We thank all the involved families for participating in and supporting this study.
Funding
This work was supported by the National Key Research and Developmental Program of China (2022YFC2702604 to Y.-Q.T.), the National Natural Science Foundation of China (81971447 and 82171608 to Y.-Q.T.,), the Hunan Provincial Natural Science Foundation of China (2023JJ40459 to W.L.W., 2022JJ30772 to Ju.D.), China Postdoctoral Science Foundation (2022M721124 to W.L.W.), and Research Grant from Reproductive and Genetic Hospital of CITIC-Xiangya (YNXM-202305 to W.L.W., YNXM-202002 to Ju.D.).
Author information
Authors and Affiliations
Contributions
Conceptualization: Ju.D., G.L., G.X.L., Y.-Q.T.; investigation: X.H., W.L.W., Ji.D.; funding acquisition: Ju.D., Y.-Q.T., W.L.W.; methodology: X.H., Y.Z., W.B.H.; clinical resources: F.G., K.L.L., S.P.Z., L.L.Y; project administration and supervision: G.L., Y.-Q.T., Q.J.Z., W.L.; validation: Ji.D., Z.X.W., Y.Z., Q.T.; writing—original draft: W.L.W., X.H.; writing—review and editing: Ju.D., G.L.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
G.L. and Ju.D. cosupervised the study and should be considered shared last authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, X., Wang, W., Luo, K. et al. Extended application of PGT-M strategies for small pathogenic CNVs. J Assist Reprod Genet 41, 739–750 (2024). https://doi.org/10.1007/s10815-024-03028-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-024-03028-6