Abstract
Background
Although the importance of miRNA variants in female reproductive disorders has been frequently reported, the association between miRNA polymorphisms and recurrent pregnancy loss (RPL) has been poorly studied. In this study, we aimed to assess the correlation of four different miRNA variants to unexplained RPL.
Methods and results
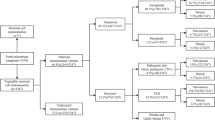
The prevalence of four SNPs including miR-21 rs1292037, miR-155-5p rs767649, miR-218–2 rs11134527, and miR-605 rs2043556 in 280 cases with iRPL and 280 controls was performed. The DNA was extracted from all subjects and the SNPs were genotyped using RFLP-PCR methods. The data revealed that rs1292037 and rs767649 were significantly associated with higher rates of iRPL in patients compared with controls while rs11134527 and rs2043556 showed no association with increased rates of iRPL among patients. The haplotypes T-A-G-G and T-A-G-A were the most frequent in both cases and controls. Three haplotypes including T-T-G-A, C-T-G-G, and T-A-A-A showed significantly different frequencies in patients in comparison to healthy females.
Conclusion
This study suggests that rs1292037 and rs767649 could be risk factors for increased rates of iRPL.
Similar content being viewed by others
Data availability
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
References
Dimitriadis E, et al. Recurrent pregnancy loss. Nat Rev Dis Primers. 2020;6(1):1–19.
Farsimadan M, et al. The effects of hepatitis B virus infection on natural and IVF pregnancy: A meta-analysis study. J Viral Hepatitis. 2021;28(9):1234–45.
Siahpoosh Z, et al. KISS1R polymorphism rs587777844 (Tyr313His) is linked to female infertility. Br J Biomed Sci. 2021;78(2):98–100.
Tara SS, et al. Methylenetetrahydrofolate Reductase C677T and A1298C polymorphisms in male partners of recurrent miscarriage couples. J Reprod Infertil. 2015;16(4):193–8.
Meng F, et al. KISS1 gene variations and susceptibility to idiopathic recurrent pregnancy loss. Reprod Sci. 2023;30:1–7.
Liu D-Y, et al. SNP rs12794714 of CYP2R1 is associated with serum vitamin D levels and recurrent spontaneous abortion (RSA): a case–control study. Arch Gynecol Obstet. 2021;304:179–90.
Rai S, et al. Correlation of follicle-stimulating hormone receptor gene Asn 680 Ser (rs6166) polymorphism with female infertility. J Family Med Prim Care. 2019;8(10):3356–61.
Farsimadan M, et al. Association analysis of KISS1 polymorphisms and haplotypes with polycystic ovary syndrome. Br J Biomed Sci. 2021;78(4):201–5.
Haqiqi H, et al. Association of FSHR missense mutations with female infertility, in silico investigation of their molecular significance and exploration of possible treatments using virtual screening and molecular dynamics. Anal Biochem. 2019;586:113433.
Jodeiryzaer S, et al. Association of oestrogen receptor alpha gene SNPs Arg157Ter C> T and Val364Glu T> A with female infertility. Br J Biomed Sci. 2020;77(4):216–8.
Wang XQ, et al. Haplotype-based association of two SNPs in miR-423 with unexplained recurrent pregnancy loss in a Chinese Han population. Exp Cell Res. 2019;374(1):210–20.
Jairajpuri DS, et al. Differentially expressed circulating microRNAs associated with idiopathic recurrent pregnancy loss. Gene. 2021;768:145334.
Ezat SA, Haji AI. Study of association between different microRNA variants and the risk of idiopathic recurrent pregnancy loss. Arch Gynecol Obstet. 2022;306(4):1281–6.
Cho SH, et al. Genetic polymorphisms in miR-604A>G, miR-938G>A, miR-1302–3C>T and the risk of idiopathic recurrent pregnancy loss. Int J Mol Sci. 2021;22(11).6127.
Tersigni C, et al. Recurrent pregnancy loss is associated to leaky gut: a novel pathogenic model of endometrium inflammation? J Transl Med. 2018;16:1–9.
Li R, et al. Genetic variants miR-126, miR-146a, miR-196a2, and miR-499 in polycystic ovary syndrome. Br J Biomed Sci. 2022;79:7.
Gomari MM, et al. CD44 polymorphisms and its variants, as an inconsistent marker in cancer investigations. Mutat Res/Rev Mutat Res. 2021;787:108374.
Santamaria X, Taylor H. MicroRNA and gynecological reproductive diseases. Fertil Steril. 2014;101(6):1545–51.
Tsai E-M, et al. A microRNA-520 mirSNP at the MMP2 gene influences susceptibility to endometriosis in Chinese women. J Hum Genet. 2013;58(4):202–9.
Jaafar S, et al. MicroRNA variants miR-27a rs895819 and miR-423 rs6505162, but not miR-124–1 rs531564, are linked to endometriosis and its severity. Br J Biomed Sci. 2022;79:9.
Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil Steril. 2020;113(3):533–5.
Antoine D, et al. Rapid, Point-of-Care scFv-SERS assay for Femtogram level detection of SARS-CoV-2. ACS Sensors. 2022;7(3):866–73.
Hocaoglu M, et al. Identification of miR-16-5p and miR-155-5p microRNAs differentially expressed in circulating leukocytes of pregnant women with polycystic ovary syndrome and gestational diabetes. Gynecol Endocrinol. 2021;37(3):216–20.
Park JH, et al. Saponin extracts induced apoptosis of endometrial cells from women with endometriosis through modulation of miR-21-5p. Reprod Sci. 2018;25:292–301.
Lee HA, et al. Association between miR-605A>G, miR-608G>C, miR-631I>D, miR-938C>T, and miR-1302-3C>T polymorphisms and risk of recurrent implantation failure. Reprod Sci. 2019;26(4):469–75.
You W, Wang Y, Zheng J. Plasma miR-127 and miR-218 might serve as potential biomarkers for cervical cancer. Reprod Sci. 2015;22(8):1037–41.
Dezfuli NK, et al. The miR-146a SNP Rs2910164 and miR-155 SNP rs767649 are risk factors for non-small cell lung cancer in the iranian population. Can Respir J. 2020;2020:8179415.
Yang J, et al. Association study of relationships of polymorphisms in the miR-21, miR-26b, miR-221/222 and miR-126 genes with cervical intraepithelial neoplasia and cervical cancer. BMC Cancer. 2021;21:1–9.
Wu Y, et al. Predictive value of miR-219–1, miR-938, miR-34b/c, and miR-218 polymorphisms for gastric cancer susceptibility and prognosis. Dis Markers. 2017;2017:4731891.
Barchitta M, et al. The role of miRNAs as biomarkers for pregnancy outcomes: a comprehensive review. Int J Genom. 2017;2017:8067972.
Yang H, et al. Clinical application of exosomes and circulating microRNAs in the diagnosis of pregnancy complications and foetal abnormalities. J Transl Med. 2020;18(1):1–9.
Ali A, et al. MicroRNA–mRNA networks in pregnancy complications: A comprehensive downstream analysis of potential biomarkers. Int J Mol Sci. 2021;22(5):2313.
Xu P, et al. Placenta-derived microRNAs in the pathophysiology of human pregnancy. Front Cell Dev Biol. 2021;9:646326.
Fu G, et al. MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci. 2013;14(3):5519–44.
Tsochandaridis M, et al. Circulating microRNAs as clinical biomarkers in the predictions of pregnancy complications. BioMed Res Int. 2015;2015:294954.
Kamalidehghan B, et al. The importance of small non-coding RNAs in human reproduction: a review article. Appl Clin Genet. 2020;13:1–11.
Li L, et al. miRNA-210-3p regulates trophoblast proliferation and invasiveness through fibroblast growth factor 1 in selective intrauterine growth restriction. J Cell Mol Med. 2019;23(6):4422–33.
Siddiqui ZH, et al. The role of miRNA in somatic embryogenesis. Genomics. 2019;111(5):1026–33.
Lv Y, et al. Roles of microRNAs in preeclampsia. J Cell Physiol. 2019;234(2):1052–61.
Liu S, et al. Identification of key circRNAs/lncRNAs/miRNAs/mRNAs and pathways in preeclampsia using bioinformatics analysis. Med Sci Monit Int Med J Exp Clin Res. 2019;25:1679.
Hornakova A, et al. Diagnostic potential of microRNAs as biomarkers in the detection of preeclampsia. Genet Test Mol Biomarkers. 2020;24(6):321–7.
Tian QX, et al. Comprehensive analysis of the differential expression profile of microRNAs in missed abortion. Kaohsiung J Med Sci. 2020;36(2):114–21.
Shahidi M, et al. miR-146b-5p and miR-520h Expressions Are Upregulated in Serum of Women with Recurrent Spontaneous Abortion. Biochem Genet. 2022;60(5):1716–32.
Cook J, et al. First trimester circulating MicroRNA biomarkers predictive of subsequent preterm delivery and cervical shortening. Sci Rep. 2019;9(1):5861.
Winger EE, et al. MicroRNAs isolated from peripheral blood in the first trimester predict spontaneous preterm birth. PLoS ONE. 2020;15(8):e0236805.
Garcia-Beltran C, et al. microRNAs in newborns with low birth weight: relation to birth size and body composition. Pediatr Res. 2022;92(3):829–37.
Wang D, et al. Altered expression of miR-518b and miR-519a in the placenta is associated with low fetal birth weight. Am J Perinatol. 2014;31(9):729–34.
Sun J, et al. Exosomal MicroRNAs in serum as potential biomarkers for ectopic pregnancy. Biomed Res Int. 2020;2020:3521859.
Kontomanolis EN, Kalagasidou S, Fasoulakis Z. MicroRNAs as potential serum biomarkers for early detection of ectopic pregnancy. Cureus. 2018;10(3):e2344.
Juchnicka I, et al. miRNAs as predictive factors in early diagnosis of gestational diabetes mellitus. Front Endocrinol (Lausanne). 2022;13:839344.
Zhang Z, et al. The possible role of visceral fat in early pregnancy as a predictor of gestational diabetes mellitus by regulating adipose-derived exosomes miRNA-148 family: protocol for a nested case-control study in a cohort study. BMC Pregnancy Childbirth. 2021;21(1):262.
Légaré C, et al. First trimester plasma MicroRNA levels predict risk of developing gestational diabetes mellitus. Front Endocrinol (Lausanne). 2022;13:928508.
Arias-Sosa LA, et al. Genetic and epigenetic variations associated with idiopathic recurrent pregnancy loss. J Assist Reprod Genet. 2018;35(3):355–66.
Ng S-W, et al. Endometrial decidualization: the primary driver of pregnancy health. Int J Mol Sci. 2020;21(11):4092.
Légaré C, et al. Human plasma pregnancy-associated miRNAs and their temporal variation within the first trimester of pregnancy. Reprod Biol Endocrinol. 2022;20(1):1–13.
Hosseini MK, et al. MicroRNA expression profiling in placenta and maternal plasma in early pregnancy loss. Mol Med Rep. 2018;17(4):4941–52.
Hayder H, et al. MicroRNAs: crucial regulators of placental development. Reproduction. 2018;155(6):R259-r271.
Xue P, et al. miR-155* mediates suppressive effect of PTEN 3′-untranslated region on AP-1/NF-κB pathway in HTR-8/SVneo cells. Placenta. 2013;34(8):650–6.
Maccani MA, Padbury JF, Marsit CJ. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS ONE. 2011;6(6):e21210.
Kochhar P, et al. Placental expression of miR-21-5p, miR-210-3p and miR-141-3p: relation to human fetoplacental growth. Eur J Clin Nutr. 2022;76(5):730–8.
Zhang JT, et al. Decreased miR-143 and increased miR-21 placental expression levels are associated with macrosomia. Mol Med Rep. 2016;13(4):3273–80.
Guan C-Y, et al. Down-regulated miR-21 in gestational diabetes mellitus placenta induces PPAR-α to inhibit cell proliferation and infiltration. Diabetes Metab Syndr Obes Targets Ther. 2020;13:3009.
El-Shorafa HM, Sharif FA. Levels of miR-21and miR-182 in unexplained recurrent spontaneous abortion. Int J Chem Lifesciences. 2019;2(6):1185–8.
Dong K, et al. Downregulations of circulating miR-31 and miR-21 are associated with preeclampsia. Pregnancy Hypertens. 2019;17:59–63.
Zhou F, et al. microRNA-21 regulates the proliferation of placental cells via FOXM1 in preeclampsia. Exp Ther Med. 2020;20(3):1871–8.
Dong K, et al. Down-regulated placental miR-21 contributes to preeclampsia through targeting RASA1. Hypertens Pregnancy. 2021;40(3):236–45.
Zhang Y, et al. MicroRNA-155 contributes to preeclampsia by down-regulating CYR61. Am J Obstet Gynecol. 2010;202(5):466.e1-7.
Li X, et al. MicroRNA-155 inhibits migration of trophoblast cells and contributes to the pathogenesis of severe preeclampsia by regulating endothelial nitric oxide synthase. Mol Med Rep. 2014;10(1):550–4.
Xu P, et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension. 2014;63(6):1276–84.
Brkić J, et al. MicroRNA-218-5p promotes endovascular trophoblast differentiation and spiral artery remodeling. Mol Ther. 2018;26(9):2189–205.
Fang M, et al. Hypoxia-inducible microRNA-218 inhibits trophoblast invasion by targeting LASP1: Implications for preeclampsia development. Int J Biochem Cell Biol. 2017;87:95–103.
Yu Z, et al. LncRNA SNHG16 regulates trophoblast functions by the miR-218-5p/LASP1 axis. J Mol Histol. 2021;52(5):1021–33.
Farsimadan M, et al. MicroRNA variants in endometriosis and its severity. Br J Biomed Sci. 2021;78(4):206–10.
Alipour M, et al. Association between miR-146a C > G, miR-149 T > C, miR-196a2 T > C, and miR-499 A > G polymorphisms and susceptibility to idiopathic recurrent pregnancy loss. J Assist Reprod Genet. 2019;36(11):2237–44.
Hu Y, et al. Functional study of one nucleotide mutation in pri-miR-125a coding region which related to recurrent pregnancy loss. PLoS ONE. 2014;9(12):e114781.
Poltronieri-Oliveira AB, et al. Polymorphisms of miR-196a2 (rs11614913) and miR-605 (rs2043556) confer susceptibility to gastric cancer. Gene Rep. 2017;7:154–63.
Zhang J, et al. Correlations of MicroRNA-21 gene polymorphisms with chemosensitivity and prognosis of cervical cancer. Am J Med Sci. 2018;356(6):544–51.
Yang J, et al. Association study of relationships of polymorphisms in the miR-21, miR-26b, miR-221/222 and miR-126 genes with cervical intraepithelial neoplasia and cervical cancer. BMC Cancer. 2021;21(1):997.
Xiang Y, et al. Association of miR-21, miR-126 and miR-605 gene polymorphisms with ischemic stroke risk. Oncotarget. 2017;8(56):95755–63.
Li H, et al. Association of genetic variants in lncRNA GAS5/miR-21/mTOR axis with risk and prognosis of coronary artery disease among a Chinese population. J Clin Lab Anal. 2020;34(10):e23430.
Ayoub SE, et al. Association of MicroRNA-155rs767649 Polymorphism with Susceptibility to Preeclampsia. Int J Mol Cell Med. 2019;8(4):247–57.
Wang S, et al. The rs767649 polymorphism in the promoter of miR-155 contributes to the decreased risk for cervical cancer in a Chinese population. Gene. 2016;595(1):109–14.
Ahmed Ali M, et al. Relationship between miR-155 and miR-146a polymorphisms and susceptibility to multiple sclerosis in an Egyptian cohort. Biomed Rep. 2020;12(5):276–84.
Assmann TS, et al. Polymorphisms in genes encoding miR-155 and miR-146a are associated with protection to type 1 diabetes mellitus. Acta Diabetol. 2017;54(5):433–41.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. This study was confirmed by the ethics review board of the Department of Obstetrics, Hengshui People’s Hospital with a full ethical code (2020–0041-25).
Consent to publish
All the women were asked to read and sign the consent form.
Conflict of interest
None.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, C., Yin, X. & Yao, S. The effect of MicroRNAs variants on idiopathic recurrent pregnancy loss. J Assist Reprod Genet 40, 1589–1595 (2023). https://doi.org/10.1007/s10815-023-02827-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02827-7