Abstract
Purpose
To analyze the influence of Caucasian mitochondrial haplogroups on controlled ovarian stimulation outcome (COS), embryo (E), and pregnancy success.
Methods
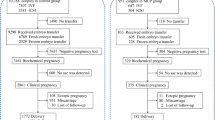
In a Caucasian population (n = 517) undergoing COS, mitochondrial haplogroups and physiological parameters were determined. Patients were classified, according to Bologna criteria, as good (>3)/poor ≤3) responder, on dependence of recruited oocytes (RO), and in pregnancy/non-pregnancy groups. Haplogroups were determined by sequencing mitochondrial hypervariable sequence I and confirmed by polymerase chain reaction (PCR), followed by restriction fragment length polymorphisms (RFLP).
Results
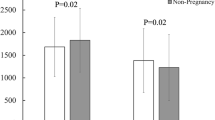
The rank of total dose of FSH (TD FSH) was similar in all clusters/haplogroups, except in JT, which is narrower (950–3,650 IU), particularly in T (1,350–3,650 IU). The statistical analysis showed higher RO and E in JT when compared to U, although it was only Uk which accumulated significantly in pregnancy respect to JT. Pearson’s correlations between TD FSH and RO showed negative statistical significance in all population (P = 0.001), H (P = 0.03), JT (P = 0.01), and T (P = 0.03). The percentage of contribution of TD FSH on RO was almost nine times in the JT cluster as compared to all population one.
Conclusions
JT cluster shows a different influence of TD FSH on RO. JT cluster shows higher RO and E than U, but it is Uk which exhibits a significant higher pregnancy rate than JT. The negative influence of the JT cluster on pregnancy success strongly suggests that the m.4216 T > C polymorphism could be responsible.


Similar content being viewed by others
Data Availability
All data are available upon request.
Web resources:
MITOMAP: A Human Mitochondrial Genome Database. http://www.mitomap.org, 2019. Statistical power of the study was determined by Epidat 3.1. http://scielo.isciii.es/scielo.php?script=sci_arttext&pid=S1135-57272004000200013&lng=es&nrm=iso>. ISSN 2173-9110.
Abbreviations
- ATP:
-
Adenosine triphosphate
- COS:
-
Controlled ovarian stimulation
- FSH:
-
Follicle stimulating hormone
- ICSI:
-
Intracytoplasmatic sperm injection
- IVF:
-
In vitro fertilization
- mtDNA:
-
Mitochondrial DNA
- OXPHOS:
-
Mitochondrial oxidative phosphorylation system
- PD:
-
Parkinson disease
- PCR:
-
Polymerase chain reaction
- RFLP:
-
Restriction fragment length polymorphism
- ROS:
-
Reactive oxygen species
- tRNA:
-
Transfer ribonucleic acid
- VO2max:
-
Maximal oxygen uptake
References
Bellver J, Donnez J. Introduction: infertility etiology and offspring health. Fertil Steril. 2019;111:1033–5.
May-Panloup P, Boucret L, De la Barca JM, Desquiret-Dumas V, Ferré-L’Hotellier V, Morinière C, et al. Ovarian ageing: the role of mitochondria in oocytes and follicles. Hum Reprod Update. 2016;22:725–43.
Rooney K, Domar A. The relationship between stress and infertility. Dialogues Clin Neurosci. 2018;20:41–7.
ESHRE Guideline Group on Ovarian Stimulation, Bosch E, Broer S, Griesinger G, Grynberg M, Humaidan P, et al. ESHRE guideline: ovarian stimulation for IVF/ICS. Hum Reprod Open. 2020;2:1–13.
Delvigne A, Rozenberg S. Epidemiology and prevention of ovarian hyperstimulation syndrome (OHSS): a review. Hum Reprod Update. 2002;8:559–77.
Kaiser UB. The pathogenesis of the ovarian hyperstimulation syndrome. N Engl J Med. 2003;349:729–32.
Achrekar S, Modi D, Desai S, Mangoli V, Mangoli R, Mahale S. Follicle-stimulating hormone receptor polymorphism (Thr307Ala) is associated with variable ovarian response and ovarian hyperstimulation syndrome in Indian women. Fertil Steril. 2009;91:2.
Čuš M, Vlaisavljević V, Repnik K, PotoČnik U, KovaČiČ B. Could polymorphisms of some hormonal receptor genes, involved in folliculogenesis help in predicting patient response to controlled ovarian stimulation? J Assist Reprod Genet. 2019;36:47–55.
De Castro F, Ruiz R, Montoro L, Pérez-Hernández D, Sánchez-Casas Padilla E, Real LM, et al. Role of follicle-stimulating hormone receptor Ser680Asn polymorphism in the efficacy of follicle-stimulating hormone. Fertil Steril. 2003;80:571–6.
Ilgaz N, Aydos OS, Karadag A, Taspinar M, Eryilmaz O, Sunguroglu A. Impact of follicle-stimulating hormone receptor variants in female infertility. J Assist Reprod Genet. 2015;32:1659–68.
Perez Mayorga M, Gromoll J, Behre HM, Gassner C, Nieschlag E, Simoni M. Ovarian response to follicle-stimulating hormone (FSH) stimulation depends on the FSH receptor genotype. J Clin Endocrinol Metab. 2000;85:3365–9.
Trevisan C, Peluso C, Cordts E, De Oliveira R, Christofolini DM, Barbosa C, Bianco B. Ala307Thr and Asn680Ser polymorphisms of FSHR gene in human reproduction outcomes. Cell Physiol Biochem. 2014;34:1527–35.
Monge-Ochoa B, Montoro L, Gil-Arribas E, Montoya J, Ruíz-Pesini E, López-Pérez MJ, et al. Variants Ala307Ala and Ser680Ser of 307 and 680 FSHr polymorphisms negatively influence on assisted reproductive techniques outcome and determine high probability of non-pregnancy in Caucasian patients. J Assist Reprod Genet. 2021;38:2769–79.
Ruíz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace D. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–6.
Rao VK, Carlson EA, Yan SS. Mitochondrial permeability transition pore is a potential drug target for neurodegeneration. Biochim Biophys Acta. 2014;1842:1267–72.
Sato M, Sato K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim Biophys Acta. 2013;1833:1979–84.
Carelli V. Keeping in shape the dogma of mitochondrial DNA maternal inheritance. PloS Genet. 2015;11:1005179.
Ruíz-Pesini E, Lott T, Procaccio V, Poole C, Marty C, Brandon M, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:823–8.
Massaad CA, Klann E. Reactive oxygen species in the regulation of synaptic plasticity and memory. Antioxid Redox Signal. 2011;14:2013–54.
Mattiazzi M, Vijayvergiya C, Gajewski CD, DeVivo DC, Lenaz G, Wiedmann M, et al. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–79.
Nijtmans LG, Henderson NS, Attardi G, Holt IJ. Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. J Biol Chem. 2001;276:6755–62.
Tranah GJ, Santaniello A, Caillier S, D’Alfonso S, Boneschi FM, et al. Mitochondrial DNA sequence variation in multiple sclerosis. Neurology. 2015;85:325–30.
Majamaa K, Finnilä S, Turkka J, Hassinen IE. Mitochondrial DNA haplogroup U as a risk factor for occipital stroke in migraine. Lancet. 1998;352:455–6.
Majamaa K, Moilanen J, Uimonen S, Remes A, Salmela P, et al. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: prevalence of the mutation in an adult population. Genet. 1998;63:447–54.
Domínguez-Garrido E, Martínez-Redondo D, Martín-Ruiz C, Gómez-Durán A, Ruíz-Pesini E, Madero P, et al. Association of mitochondrial haplogroup J and mtDNA oxidative damage in two different North Spain elderly populations. Biogerontology. 2009;10:435–42.
Montiel-Sosa F, Ruiz-Pesini E, Enriquez JA, Marcuello A, Díez-Sánchez C, Montoya J, et al. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene. 2016;368:21–7.
Ruíz-Pesini E, Lapeña AC, Díez-Sánchez C, Pérez-Martos A, Montoya J, Álvarez E, et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet. 2000;67:682–96.
Marcuello A, Martínez-Redondo D, Dahmani Y, Casajús JA, Ruiz-Pesini E, Montoya J, et al. Human mitochondrial variants influence on oxygen consumption. Mitochondrion. 2009;9:27–30.
Otten A, Smeets H. Evolutionary defined role of the mitochondrial DNA in fertility, disease and ageing. Hum Reprod Update. 2015;21:671–89.
Danhua P, Wu J, Liu J. Comparisons of GnRH antagonist versus GnRH agonist protocol in poor ovarian responders undergoing IVF. Human Reprod. 2011;26:2742–9.
Demirol A, Gurgan T. Comparison of microdose flare-up and antagonist multiple-dose protocols for poor-responder patients: a randomized study. Fertil Steril. 2009;92:481–5.
Griesinger G, Diedrich K, Devroey P, Kolibianakis EM. GnRH agonist for triggering final oocyte maturation in the GnRH antagonist ovarian hyperstimulation protocol: a systematic review and meta-analysis. Human Reprod Update. 2006;12:159–68.
Ferraretti AP, Goossens V, Kupka M, Bhattacharya S, De Mouzon J, Castilla JA, et al. European IVF-monitoring (EIM) consortium for the European Society of Human Reproduction and Embryology (ESHRE). Assisted reproductive technology in Europe, 2009: results generated from European registers by ESHRE. Human Reprod. 2013;28:2318–31.
Criterios ASEBIR de Valoración Morfológica de Oocitos. Embriones Tempranos y Blastocistos Humanos. In: Revista ASEBIR. 3rd ed; 2015.
Løssl K, Oldenburg A, Toftager M, Bogstad J, Praetorius L, Zedeler A, et al. Predictive value of plasma human chorionic gonadotropin measured 14 days after day-2 single embryo transfer. Acta Obstet Gynecol Scand. 2017;96:960–7.
Abu-Musa A, Haahr T, Humaidan P. Novel physiology and definition of poor ovarian response; clinical recommendations. Int J Mol Sci. 2020;21:2110.
Drakopoulos P, Blockeel C, Stoop D, Camus M, de Vos M, Tournaye H, et al. Conventional ovarian stimulation and single embryo transfer for IVF/ICSI. How many oocytes do we need to maximize cumulative live birth rates after utilization of all fresh and frozen embryos? Hum Reprod. 2016;31:370–6.
Ferraretti AP, La Marca A, Fauser BC, Tarlatzis B, Nargund G, Gianaroli L. On behalf of the ESHRE working group on poor ovarian response definition. ESHRE consensus on the definition of “poor response” to ovarian stimulation for in vitro fertilization: the Bologna criteria. Hum Reprod. 2011;26:1616–24.
Ferraretti AP, Gianaroli L. The Bologna criteria for the definition of poor ovarian responders: is there a need for revision? Hum Reprod. 2014;29:1842–5.
Patrizio P, Vaiarelli A, Setti P, Tobler K, Shoham G, Leong M, et al. How to define, diagnose and treat poor responders? Responses from a worldwide survey of IVF clinics. Reprod Biomed Online. 2015;30:581–92.
Weitzman VN, Engmann L, DiLuigi A, Maier D, Nulsen J, Benadiva C. Comparison of luteal estradiol patch and gonadotropin-releasing hormone antagonist suppression protocol before gonadotropin stimulation versus microdose gonadotropin-releasing hormone agonist protocol for patients with a history of poor in vitro fertilization outcomes. Fertil Steril. 2009;92:226–30.
Macaulay V, Richards M, Sykes B. Mitochondrial DNA recombination-no need to panic. Proc Biol Sci. 1999;266:2037–9.
Kivisild T, Shen P, Wall D, Do B, Sung R, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–87.
Muñiz-García J, Santiago-Pérez MI. ¿ Cuántos pacientes selecciono para mi estudio? Angiología. 2006;58:145–50.
DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am J Med Genet. 2001;106:18–26.
Vanniarajan A, Govindaraj P, Carlus SJ, Aruna M, Aruna P, Kumar A, et al. Mitochondrial DNA variations associated with recurrent pregnancy loss among Indian women. Mitochondrion. 2011;11:450–6.
Colagar A, Mosaieby E, Seyedhassani S, Mohajerani M, Arasteh A, Kamalidehghan B, et al. T4216C mutation in NADH dehydrogenase I gene is associated with recurrent pregnancy loss. Mitochondrial DNA. 2013;24:610–2.
Ali Azadi A, Joo Seo D, Jafari Sasansara H, Van Haute M. Mitochondrial DNA variations are associated with recurrent pregnancy loss. Mitochondrial DNA A. 2018;29:674–8.
Van der Walt J, Nicodemus K, Martin E, Scott W, Nance M, Watts R, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–11.
Fernández E, García-Moreno JM, Pablos AM, Chacón J. May the evaluation of nitrosative stress through selective increase of 3-nitrotyrosine proteins other than nitroalbumin and dominant tyrosine-125/136 nitrosylation of serum α-synuclein serve for diagnosis of sporadic Parkinson's disease? Antioxid Redox Signal. 2013;19:912–8.
Jenner P. Oxidative stress in Parkinson's disease. Ann Neurol. 2013;53:26–36.
Acknowledgements
The authors would like to express their gratitude to all patients and to their partners, who contributed genetic material for this study.
Funding
BM-O was supported by a grant of the Foundation for Biomedical Research of the Hospital Universitario Príncipe de Asturias of Alcalá de Henares, Madrid (Spain). The study was supported by a grant from the Government of Aragon (Spain) (Applied Research Group B33 of 2009. Main Researcher Dr. Julio Montoya). The funding sources had no involvement in study design, collection, analysis, interpretation of data, writing the report or decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
BM-O performed the laboratory work of the study and contributed to writing the manuscript information. FC and LM collected data and provided subjects’ treatment. ML-P, JM, and ER-P did the laboratory studies for the mitochondrial DNA haplogroups determination. ML-P and FC designed the study. BM-O and CD-S performed statistical analysis of data. ML-P and CD-S supervised quality control of molecular biology studies and designed RFLP studies. CD-S contributed to study design and wrote the manuscript information.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The subjects were fully informed of the aims of the work before signing the informed consent form. The study conforms to the code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved (January 22, 2003) by the Committee of Clinical Trials of the Hospital Universitario Príncipe de Asturias of Alcalá de Henares, Madrid (Spain).
Consent for publication
All authors agree in publishing the data contained in this article.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Manuel J. López-Pérez and Francisco de Castro are deceased.
Supplementary information

Supplementary Figure 1
Phylogenetic tree of mtDNA clusters and haplogroups. Changes respect the Revised Cambridge Reference Sequence (rCRS) of the Human Mitochondrial DNA. The corresponding polymorphism is indicated in some haplogroups and subhaplogroups with the restriction enzyme and the Restriction Fragment Length Polymorphisms (RFLP) used for their identification. In this figure, the most important Caucasian variants can be observed. From left to right: cluster U and, in particular, U1811 (framed), cluster JT (framed) characterized by the polymorphism m.4216 T > C and cluster H/HV (framed). The set of these mitochondrial variants adds up more than 90% of the Caucasian population. The rest would correspond to IWX and those who cannot be ascribed to any of these variants. Figure modified from [18]. (PNG 462 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Monge-Ochoa, B., Montoro, L., Montoya, J. et al. m.4216 T > C polymorphism in JT cluster determines a lower pregnancy rate in response to controlled ovarian stimulation treatment. J Assist Reprod Genet 40, 671–682 (2023). https://doi.org/10.1007/s10815-023-02721-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02721-2