Abstract
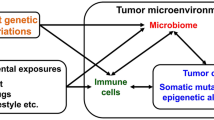
Colorectal cancer (CRC) is a leading cause of cancer-related deaths in both the USA and the world. Recent research has demonstrated the involvement of the gut microbiota in CRC development and progression. Microbial biomarkers of disease have focused primarily on the bacterial component of the microbiome; however, the viral portion of the microbiome, consisting of both bacteriophages and eukaryotic viruses, together known as the virome, has been lesser studied. Here we review the recent advancements in high-throughput sequencing (HTS) technologies and bioinformatics, which have enabled scientists to better understand how viruses might influence the development of colorectal cancer. We discuss the contemporary findings revealing modulations in the virome and their correlation with CRC development and progression. While a variety of challenges still face viral HTS detection in clinical specimens, we consider herein numerous next steps for future basic and clinical research. Clinicians need to move away from a single infectious agent model for disease etiology by grasping new, more encompassing etiological paradigms, in which communities of various microbial components interact with each other and the host. The reporting and indexing of patient health information, socioeconomic data, and other relevant metadata will enable identification of predictive variables and covariates of viral presence and CRC development. Altogether, the virome has a more profound role in carcinogenesis and cancer progression than once thought, and viruses, specific for either human cells or bacteria, are clinically relevant in understanding CRC pathology, patient prognosis, and treatment development.

Similar content being viewed by others
References
Pan D, Nolan J, Williams KH, et al. Abundance and distribution of microbial cells and viruses in an alluvial aquifer. Front Microbiol. 2017;8:1199.
Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol. 2009;11:2771–2774.
Navarro F, Muniesa M. Phages in the Human Body. Front Microbiol. 2017;8:566.
Bekliz M, Colson P, La Scola B. The expanding family of virophages. Viruses. 2016;8:317.
Caputo M, Zoch-Lesniak B, Karch A, et al. Bacterial community structure and effects of picornavirus infection on the anterior nares microbiome in early childhood. BMC Microbiol. 2019;19:1. https://doi.org/10.1186/s12866-018-1372-8.
Yinda CK, Vanhulle E, Conceição-Neto N, et al. Gut virome analysis of cameroonians reveals high diversity of enteric viruses, including potential interspecies transmitted viruses. mSphere. 2019;4:e00585-18. https://doi.org/10.1128/mSphere.00585-18.
Ly M, Abeles SR, Boehm TK, et al. Altered oral viral ecology in association with periodontal disease. mBio. 2014;5:1133. https://doi.org/10.1128/mbio.01133-14.
Ungaro F, Massimino L, Furfaro F, et al. Metagenomic analysis of intestinal mucosa revealed a specific eukaryotic gut virome signature in early-diagnosed inflammatory bowel disease. Gut Microbes. 2019;10:149–158. https://doi.org/10.1080/19490976.2018.1511664.
Fernandes MA, Verstraete SG, Phan TG, et al. Enteric virome and bacterial microbiota in children with ulcerative colitis and crohn disease. J Pediatr Gastroenterol Nutr. 2019;68:30–36. https://doi.org/10.1097/MPG.0000000000002140.
Willner D, Furlan M, Haynes M, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. 2009;4:e7370. https://doi.org/10.1371/journal.pone.0007370.
Moran Losada P, Chouvarine P, Dorda M, et al. The cystic fibrosis lower airways microbial metagenome. ERJ Open Res. 2016;2:00096–02015. https://doi.org/10.1183/23120541.00096-2015.
Santiago-Rodriguez TM, Hollister EB. Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses. 2019;11:E656.
Plummer M, de Martel C, Vignat J, et al. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609–e616.
Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674.
Monaco CL, Gootenberg DB, Zhao G, et al. Altered virome and bacterial microbiome in human immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe. 2016;19:311–322. https://doi.org/10.1016/j.chom.2016.02.011.
Abeles SR, Ly M, Santiago-Rodriguez TM, et al. Effects of long term antibiotic therapy on human oral and fecal viromes. PLoS ONE. 2015;10:e0134941.
Santiago-Rodriguez TM, Ly M, Bonilla N, et al. The human urine virome in association with urinary tract infections. Front Microbiol. 2015;6:14.
Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. https://doi.org/10.1016/j.cell.2015.01.002.
Bzhalava D, Guan P, Franceschi S, et al. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology. 2013;445:224–231.
Braaten KP, Laufer MR. Human papillomavirus (HPV), HPV-related disease, and the HPV vaccine. Rev Obstet Gynecol. 2008;1:2.
Kreimer AR, Clifford GM, Boyle P, et al. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomark Prev. 2005;14:467–475.
D’Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–1956.
Muñoz N, Bosch X, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527.
Yin B, Liu W, Yu P, et al. Association between human papillomavirus and prostate cancer: a meta-analysis. Oncol Lett. 2017;14:1855–1865. https://doi.org/10.3892/ol.2017.6367.
Damin DC, Ziegelmann PK, Damin AP. Human papillomavirus infection and colorectal cancer risk: a meta-analysis. Colorectal Dis. 2013;15:e420–e428.
Araldi RP, Sant’Ana TA, Módolo DG, et al. The human papillomavirus (HPV)-related cancer biology: an overview. Biomed Pharmacother. 2018;106:1537–1556.
Lui RN, Tsoi KKF, Ho JMW, et al. Global increasing incidence of young-onset colorectal cancer across 5 continents: a joinpoint regression analysis of 1,922,167 cases. Cancer Epidemiol Biomark Prev. 2019;28:1275–1282.
Doorbar J, Egawa N, Griffin H, et al. Human papillomavirus molecular biology and disease association. Rev Med Virol.. 2015;25:2–23.
Bodaghi S. Colorectal papillomavirus infection in patients with colorectal cancer. Clin Cancer Res. 2005;11:2862–2867.
De Gascun CF, Carr MJ. Human polyomavirus reactivation: disease pathogenesis and treatment approaches. Clin Dev Immunol. 2013;2013:373579.
Prado JCM, Monezi TA, Amorim AT, et al. Human polyomaviruses and cancer: an overview. Clinics (Sao Paulo). 2018;73:e558s. https://doi.org/10.6061/clinics/2018/e558s.
Dalianis T, Hirsch HH. Human polyomaviruses in disease and cancer. Virology. 2013;437:63–72.
Vilchez RA, Butel JS. Emergent human pathogen simian virus 40 and its role in cancer. Clin Microbiol Rev. 2004;17:495–508.
Khabaz MN, Nedjadi T, Gari MA, et al. Simian virus 40 is not likely involved in the development of colorectal adenocarcinoma. Future Virol. 2016;11:175–180.
Rollison DE. JC virus infection. J Clin Gastroenterol. 2010;. https://doi.org/10.1097/mcg.0b013e3181e0084b.
Khabaz MN, Nedjadi T, Garri MA, et al. BK polyomavirus association with colorectal cancer development. Genet Mol Res. 2016. https://doi.org/10.4238/gmr.15027841.
Cohen LJ. Phages trump bacteria in immune interactions. Sci Transl Med. 2019;11:eaaw5331.
Hannigan GD, Duhaime MB, Ruffin MT 4th, et al. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio. 2018;9:e02248-18.
Nakatsu G, Zhou H, Wu WKK, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155:e5.
Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6:e16393. https://doi.org/10.1371/journal.pone.0016393.
Tilg H, Adolph TE, Gerner RR, et al. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33:954–964.
Schieffer KM, Wright JR, Harris LR, et al. NOD2 genetic variants predispose one of two familial adenomatous polyposis siblings to pouchitis through microbiome dysbiosis. J Crohns Colitis. 2017;11:1393–1397. https://doi.org/10.1093/ecco-jcc/jjx083.
Gogokhia L, Buhrke K, Bell R, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019;25:e8.
Sze MA, Baxter NT, Ruffin MT 4th, et al. Normalization of the microbiota in patients after treatment for colonic lesions. Microbiome. 2017;5:150.
Baxter NT, Ruffin MT 4th, Rogers MAM, et al. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8:37.
Baxter NT, Koumpouras CC, Rogers MAM, et al. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome. 2016;4:59.
Zackular JP, Rogers MAM, Ruffin MT 4th, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res. 2014;7:1112–1121.
Geuking MB, Weber J, Dewannieux M, et al. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science. 2009;323:393–396.
Horie M, Honda T, Suzuki Y, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87.
Klenerman P, Hengartner H, Zinkernagel RM. A non-retroviral RNA virus persists in DNA form. Nature. 1997;390:298–301.
Zhdanov VM. Integration of viral genomes. Nature. 1975;256:471–473.
Enam S, del Valle L, Lara C, et al. Association of human polyomavirus JCV with colon cancer: evidence for interaction of viral T-antigen and beta-catenin. Cancer Res. 2002;62:7093–7101.
Goel A, Li MS, Nagasaka T, et al. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology. 2006;130:1950–1961.
Hori R, Murai Y, Tsuneyama K, et al. Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Arch. 2005;447:723–730.
Karpinski P, Myszka A, Ramsey D, et al. Detection of viral DNA sequences in sporadic colorectal cancers in relation to CpG island methylation and methylator phenotype. Tumour Biol.. 2011;32:653–659.
Mou X, Chen L, Liu F, et al. Prevalence of JC virus in Chinese patients with colorectal cancer. PLoS ONE. 2012;7:e35900. https://doi.org/10.1371/journal.pone.0035900.
Ouaïssi M, Studer AS, Mege D, et al. Characteristics and natural history of patients with colorectal cancer complicated by infectious endocarditis. Case control study of 25 patients. Anticancer Res. 2014;34:349–353.
Theodoropoulos G, Panoussopoulos D, Papaconstantinou I, et al. Assessment of JC polyoma virus in colon neoplasms. Dis Colon Rectum. 2005;48:86–91.
Vilkin A, Ronen Z, Levi Z, et al. Presence of JC virus DNA in the tumor tissue and normal mucosa of patients with sporadic colorectal cancer (CRC) or with positive family history and Bethesda criteria. Dig Dis Sci. 2012;57:79–84. https://doi.org/10.1007/s10620-011-1855-z.
zur Hausen H. Red meat consumption and cancer: reasons to suspect involvement of bovine infectious factors in colorectal cancer. Int J Cancer. 2012;130:2475–2483.
Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208–215. https://doi.org/10.1093/cid/ciu787.
Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. https://doi.org/10.1101/gr.126516.111.
Cuevas-Ramos G, Petit CR, Marcq I, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA. 2010;107:11537–11542. https://doi.org/10.1073/pnas.1001261107.
Niu YD, McAllister TA, Nash JHE, et al. Four Escherichia coli O157:H7 phages: a new bacteriophage genus and taxonomic classification of T1-like phages. PLoS ONE. 2014;9:e100426.
Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA. 2014;111:18321–18326. https://doi.org/10.1073/pnas.1406199111.
Johnson CH, Dejea CM, Edler D, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891–897. https://doi.org/10.1016/j.cmet.2015.04.011.
Secor PR, Sweere JM, Michaels LA, et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe. 2015;18:549–559. https://doi.org/10.1016/j.chom.2015.10.013.
Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914.
Nguyen S, Baker K, Padman BS, et al. Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. mBio. 2017;8:e01874-17.
Lehti TA, Pajunen MI, Skog MS, et al. Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat Commun. 2017;8:1915.
Ogilvie LA, Jones BV. The human gut virome: a multifaceted majority. Front Microbiol. 2015;6:918.
Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169.
Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394.
Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510.
García-Arroyo L, Prim N, Martí N, et al. Benefits and drawbacks of molecular techniques for diagnosis of viral respiratory infections. Experience with two multiplex PCR assays. J Med Virol. 2016;88:45–50.
Reijans M, Dingemans G, Klaassen CH, et al. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol. 2008;46:1232–1240. https://doi.org/10.1128/JCM.02294-07.
Afshar RM, Mollaie HR. Detection of HBV resistance to lamivudine in patients with chronic hepatitis B using Zip nucleic acid probes in Kerman, southeast of Iran. Asian Pac J Cancer Prev. 2012;13:3657–3661.
Mercier-Delarue S, Vray M, Plantier JC, et al. Higher specificity of nucleic acid sequence-based amplification isothermal technology than of real-time PCR for quantification of HIV-1 RNA on dried blood spots. J Clin Microbiol. 2014;52:52–56. https://doi.org/10.1128/JCM.01848-13.
Wu D, Liu F, Liu H, et al. Detection of serum HCV RNA in patients with chronic hepatitis C by transcription mediated amplification and real-time reverse transcription polymerase chain reaction. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2014;39:664–672.
Wylie TN, Wylie KM, Herter BN, et al. Enhanced virome sequencing using targeted sequence capture. Genome Res. 2015;25:1910–1920.
Moustafa A, Xie C, Kirkness E, et al. The blood DNA virome in 8,000 humans. PLoS Pathog. 2017;13:e1006292. https://doi.org/10.1371/journal.ppat.1006292.
Kleiner M, Hooper LV, Duerkop BA. Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genom. 2015;16:7.
Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Thurber RV, Haynes M, Breitbart M, et al. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483.
Reyes A, Semenkovich NP, Whiteson K, et al. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–617.
Kim K-H, Chang H-W, Nam Y-D, et al. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol. 2008;74:5975–5985.
Wang D, Urisman A, Liu YT, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. https://doi.org/10.1371/journal.pbio.0000002.
Abbas AA, Diamond JM, Chehoud C, et al. The perioperative lung transplant virome: torque teno viruses are elevated in donor lungs and show divergent dynamics in primary graft dysfunction. Am J Transplant. 2017;17:1313–1324. https://doi.org/10.1111/ajt.14076.
Breitbart M, Hewson I, Felts B, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. https://doi.org/10.1128/jb.185.20.6220-6223.2003.
Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. https://doi.org/10.1038/nature09199.
Kim M-S, Park E-J, Roh SW, et al. Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol. 2011;77:8062–8070.
Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. https://doi.org/10.1101/gr.122705.111.
Handley SA, Devkota S. Going viral: a novel role for bacteriophage in colorectal cancer. mBio. 2019;10:e02626-18.
Zheng T, Li J, Ni Y, et al. Mining, analyzing, and integrating viral signals from metagenomic data. Microbiome. 2019;7:42. https://doi.org/10.1186/s40168-019-0657-y.
Mollerup S, Asplund M, Friis-Nielsen J, et al. High-throughput sequencing-based investigation of viruses in human cancers by multienrichment approach. J Infect Dis. 2019;220:1312–1324. https://doi.org/10.1093/infdis/jiz318.
Vinner L, Mourier T, Friis-Nielsen J, et al. Investigation of human cancers for retrovirus by low-stringency target enrichment and high-throughput sequencing. Sci Rep. 2015;5:13201. https://doi.org/10.1038/srep13201.
Mühlemann B, Jones TC, de Barros Damgaard P, et al. Ancient hepatitis B viruses from the Bronze Age to the Medieval period. Nature. 2018;557:418–423.
Hansen TA, Fridholm H, Frøslev TG, et al. New type of papillomavirus and novel circular single stranded DNA virus discovered in urban Rattus norvegicus using circular DNA enrichment and metagenomics. PLoS ONE. 2015;10:e0141952. https://doi.org/10.1371/journal.pone.0141952.
Nooij S, Schmitz D, Vennema H, et al. Overview of virus metagenomic classification methods and their biological applications. Front. Microbiol. 2018;9:749.
Posada-Cespedes S, Seifert D, Beerenwinkel N. Recent advances in inferring viral diversity from high-throughput sequencing data. Virus Res. 2017;239:17–32.
Rose R, Constantinides B, Tapinos A, et al. Challenges in the analysis of viral metagenomes. Virus Evol. 2016;2:vew022.
Wommack KE, Bhavsar J, Polson SW, et al. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand Genom Sci. 2012;6:427–439. https://doi.org/10.4056/sigs.2945050.
Roux S, Tournayre J, Mahul A, et al. Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinform. 2014;15:76.
Scheuch M, Höper D, Beer M. RIEMS: a software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinform. 2015;16:69.
Norling M, Karlsson-Lindsjö OE, Gourlé H, et al. MetLab: an in silico experimental design, simulation and analysis tool for viral metagenomics studies. PLoS ONE. 2016;11:e0160334.
Bhaduri A, Qu K, Lee CS, et al. Rapid identification of non-human sequences in high-throughput sequencing datasets. Bioinformatics. 2012;28:1174–1175.
Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46.
Lefkowitz EJ, Dempsey DM, Hendrickson RC, et al. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucl Acids Res. 2018;46:D708–D717. https://doi.org/10.1093/nar/gkx932.
Aiewsakun P, Adriaenssens EM, Lavigne R, et al. Evaluation of the genomic diversity of viruses infecting bacteria, archaea and eukaryotes using a common bioinformatic platform: steps towards a unified taxonomy. J Gen Virol. 2018;99:1331–1343.
Aiewsakun P, Simmonds P. The genomic underpinnings of eukaryotic virus taxonomy: creating a sequence-based framework for family-level virus classification. Microbiome. 2018;6:38.
Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl Environ Microbiol. 2008;74:1453–1463.
Wooley JC, Ye Y. Metagenomics: facts and artifacts, and computational challenges*. J Comput Sci Technol. 2009;25:71–81.
Tang P, Chiu C. Metagenomics for the discovery of novel human viruses. Future Microbiol. 2010;5:177–189.
Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667.
Fancello L, Raoult D, Desnues C. Computational tools for viral metagenomics and their application in clinical research. Virology. 2012;434:162–174.
Thomas T, Gilbert J, Meyer F. Metagenomics—A guide from sampling to data analysis. Microb Inform Exp. 2012;2:3.
Pallen MJ. Diagnostic metagenomics: potential applications to bacterial, viral and parasitic infections. Parasitology. 2014;141:1856–1862.
Hall RJ, Draper JL, Nielsen FGG, et al. Beyond research: a primer for considerations on using viral metagenomics in the field and clinic. Front Microbiol. 2015;6:224.
McIntyre ABR, Ounit R, Afshinnekoo E, et al. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 2017;18:182.
Nieuwenhuijse DF, Koopmans MPG. Metagenomic sequencing for surveillance of food- and waterborne viral diseases. Front Microbiol. 2017;8:230.
Randle-Boggis RJ, Helgason T, Sapp M, et al. Evaluating techniques for metagenome annotation using simulated sequence data. FEMS Microbiol Ecol. 2016;92:fiw095.
Lindgreen S, Adair KL, Gardner PP. An evaluation of the accuracy and speed of metagenome analysis tools. Sci Rep. 2016;6:19233.
Treangen TJ, Koren S, Sommer DD, et al. MetAMOS: a modular and open source metagenomic assembly and analysis pipeline. Genome Biol. 2013;14:R2.
Scholz M, Lo C-C, Chain PSG. Improved assemblies using a source-agnostic pipeline for MetaGenomic Assembly by Merging (MeGAMerge) of contigs. Sci Rep. 2015;4:6480.
Smits SL, Bodewes R, Ruiz-Gonzalez A, et al. Assembly of viral genomes from metagenomes. Front Microbiol. 2014;5:714. https://doi.org/10.3389/fmicb.2014.00714.
Vázquez-Castellanos JF, García-López R, Pérez-Brocal V, et al. Comparison of different assembly and annotation tools on analysis of simulated viral metagenomic communities in the gut. BMC Genom. 2014;15:37.
Deng X, Naccache SN, Ng T, et al. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res. 2015;43:e46. https://doi.org/10.1093/nar/gkv002.
Henry VJ, Bandrowski AE, Pepin A-S, Gonzalez BJ, Desfeux A. OMICtools: an informative directory for multi-omic data analysis. Database. 2014.
Fredericks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33.
Arias M, Fan H. The saga of XMRV: a virus that infects human cells but is not a human virus. Emerg Microbes Infect. 2014;3:1–6.
Alter HJ, Mikovits JA, Switzer WM, et al. A multicenter blinded analysis indicates no association between chronic fatigue syndrome/myalgic encephalomyelitis and either xenotropic murine leukemia virus-related virus or polytropic murine leukemia virus. mBio. 2012;3:e00266-12. https://doi.org/10.1128/mbio.00266-12.
Erlwein O, et al. DNA extraction columns contaminated with murine sequences. PLoS ONE. 2011;6:e23484.
Naccache SN, Greninger AL, Lee D, et al. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol. 2013;87:11966–11977. https://doi.org/10.1128/JVI.02323-13.
Lewandowska DW, Zagordi O, Abinden A, et al. Unbiased metagenomic sequencing complements specific routine diagnostic methods and increases chances to detect rare viral strains. Diagn Microbiol Infect Dis. 2015;83:133–138.
Schlaberg R, Chiu CY, Miller S, et al. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–786.
Kim M-S, Bae J-W. Spatial disturbances in altered mucosal and luminal gut viromes of diet-induced obese mice. Environ Microbiol. 2016;18:1498–1510.
Minot S, Bryson A, Chehoud C, et al. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450–12455. https://doi.org/10.1073/pnas.1300833110.
Nagata N, Tohya M, Fukuda S, et al. Effects of bowel preparation on the human gut microbiome and metabolome. Sci Rep. 2019;9:4042. https://doi.org/10.1038/s41598-019-40182-9.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Regina Lamendella receives funding from the National Science Foundation (NSF1805549) and the National Institute of Health - NIA (1R15AG052933-01). She is also the CEO of Wright Labs, LLC and the VP of research and development for Contamination Source Identification.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Emlet, C., Ruffin, M. & Lamendella, R. Enteric Virome and Carcinogenesis in the Gut. Dig Dis Sci 65, 852–864 (2020). https://doi.org/10.1007/s10620-020-06126-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06126-4