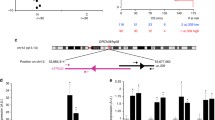
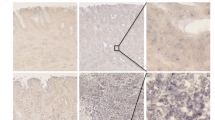
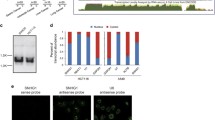
Abstract
Long noncoding RNAs have recently emerged as significant contributors to cancers, including breast cancer (BC). One class of long noncoding RNAs called transcribed ultraconserved regions (T-UCRs) is highly conserved in many species and closely related to diverse physiological and pathological processes. However, the function of T-UCRs in BC remains largely unclear. In this study, we identified uc.51, a T-UCR that is overexpressed in both BC tissues and cell lines and is correlated with larger tumor size. Loss- and gain-of-function assays were performed in vitro and demonstrated that uc.51 promotes the proliferation, migration, and invasion of BC cells. Mechanistically, non-POU domain-containing octamer-binding protein (NONO) was found to physically interact with uc.51 by RNA pulldown followed by mass spectrometry. This interaction was further verified by RNA immunoprecipitation. Moreover, uc.51 positively regulated the expression of NONO, maintained its stability through the ubiquitin–proteasome system, and activated the phosphorylation of CREB. Rescue experiments demonstrated that NONO overexpression compensated for the attenuated influence on BC progression resulting from downregulation of uc.51, indicating that NONO functions downstream of uc.51. In vivo functional experiments also revealed a positive correlation between uc.51 expression and tumor size. Ki-67 and NONO levels in the lv-uc.51-shRNA group were decreased compared with those in the lv-con-shRNA group, according to the immunohistochemical staining results, and a decreased incidence of distant metastasis was observed in the lv-uc.51-shRNA group in the xenograft model. Collectively, our results reveal a substantial role for the uc.51-NONO axis in BC progression and indicate that the uc.51-NONO axis has potential to be a therapeutic target for BC.









Similar content being viewed by others
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- lncRNAs:
-
Long noncoding RNAs
- BC:
-
Breast cancer
- T-UCRs:
-
Transcribed ultraconserved regions
- NONO:
-
Non-POU domain-containing octamer-binding protein
- RIP:
-
RNA immunoprecipitation
- ATCC:
-
American type culture collection
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- FBS:
-
Fetal bovine serum
- siRNAs:
-
Small interfering RNAs
- shRNA:
-
Short hairpin RNA
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- cDNA:
-
Complementary DNA
- FISH:
-
Fluorescence in situ hybridization
- CCK-8:
-
Cell Counting Kit-8
- PBS:
-
Phosphate buffered saline
- TBST:
-
Tris-buffered saline plus Tween
- SDS:
-
Sodium dodecyl sulfate
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PVDF:
-
Polyvinylidene fluoride
- PBS:
-
Phosphate buffered solution
- HRP:
-
Horseradish peroxidase
- Co-IP:
-
Co-immuoprecipitation
- IHC:
-
Immunohistochemistry
- H&E:
-
Hematoxylin and eosin
- RBP:
-
RNA-binding protein
References
Cao W, Chen HD, Yu YW, Li N, Chen WQ (2021) Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 134:783–791. https://doi.org/10.1097/CM9.0000000000001474
Harbeck N, Penault-Llorca F, Cortes J et al (2019) Breast cancer. Nat Rev Dis Primers 5:66. https://doi.org/10.1038/s41572-019-0111-2
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics. CA Cancer J Clin 71:7–33. https://doi.org/10.3322/caac.21654
Steeg PS (2006) Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 12:895–904. https://doi.org/10.1038/nm1469
Niu Y, Bao L, Chen Y et al (2020) HIF2-induced long noncoding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res 80:964–975. https://doi.org/10.1158/0008-5472.CAN-19-1532
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641. https://doi.org/10.1016/j.cell.2009.02.006
Schmitz SU, Grote P, Herrmann BG (2016) Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 73:2491–2509. https://doi.org/10.1007/s00018-016-2174-5
Chen LL (2016) Linking long noncoding RNA localization and function. Trends Biochem Sci 41:761–772. https://doi.org/10.1016/j.tibs.2016.07.003
Bhan A, Soleimani M, Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77:3965–3981. https://doi.org/10.1158/0008-5472.CAN-16-2634
Wang Z, Yang B, Zhang M et al (2018) lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 33:706-720.e9. https://doi.org/10.1016/j.ccell.2018.03.006
Hu YP, Jin YP, Wu XS et al (2019) LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol Cancer 18:167. https://doi.org/10.1186/s12943-019-1097-9
Wang D, Chen Z, Xu H, He A, Liu Y, Huang W (2017) Long noncoding RNA CCAT2 as a novel biomaker of metastasis and prognosis in human cancer: a meta-analysis. Oncotarget 8:75664–75674
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12:861–874. https://doi.org/10.1038/nrg3074
Bejerano G, Pheasant M, Makunin I et al (2004) Ultraconserved elements in the human genome. Science 304:1321–1325. https://doi.org/10.1126/science.1098119
Pereira Zambalde E, Mathias C, Rodrigues AC, de Souza Fonseca Ribeiro EM, Fiori-Gradia D, Calin GA, Carvalho de Oliveira J (2020) Highlighting transcribed ultraconserved regions in human diseases. Wiley Interdiscip Rev RNA 11:e1567. https://doi.org/10.1002/wrna.1567
Dickel DE, Ypsilanti AR, Pla R et al (2018) Ultraconserved enhancers are required for normal development. Cell 172:491-499.e15. https://doi.org/10.1016/j.cell.2017.12.017
Sun Y, Fan W, Xue R et al (2020) Transcribed ultraconserved regions, Uc.323, Ameliorates Cardiac Hypertrophy by Regulating the Transcription of CPT1b (Carnitine Palmitoyl transferase 1b). Hypertension 75:79–90. https://doi.org/10.1161/HYPERTENSIONAHA.119.13173
Zhang C, Peng Y, Wang Y, Xu H, Zhou X (2020) Transcribed ultraconserved noncoding RNA uc.153 is a new player in neuropathic pain. Pain 161:1744–1754. https://doi.org/10.1097/j.pain.0000000000001868
Dong B, Horowitz DS, Kobayashi R, Krainer AR (1993) Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res 21:4085–4092. https://doi.org/10.1093/nar/21.17.4085,PMID8371983
Feng PF, Li L, Deng T et al (2020) NONO and tumorigenesis: more than splicing. J Cell Mol Med 24:4368–4376. https://doi.org/10.1111/jcmm.15141
Kowalska E, Ripperger JA, Muheim C et al (2012) Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol Cell Biol 32:4585–4594. https://doi.org/10.1128/MCB.00334-12,PMID22966205
Andersen JS, Lyon CE, Fox AH et al (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12:1–11. https://doi.org/10.1016/s0960-9822(01)00650-9
Hata K, Nishimura R, Muramatsu S et al (2008) Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J Clin Invest 118:3098–3108. https://doi.org/10.1172/JCI31373
Kameoka S, Duque P, Konarska MM (2004) P54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J 23:1782–1791. https://doi.org/10.1038/sj.emboj.7600187
Krietsch J, Caron MC, Gagné JP et al (2012) PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res 40:10287–10301. https://doi.org/10.1093/nar/gks798
Yu T, Zhao Y, Hu Z et al (2017) MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res 77:5782–5794. https://doi.org/10.1158/0008-5472.CAN-17-0671
Schiffner S, Zimara N, Schmid R, Bosserhoff AK (2011) p54(nrb) is a new regulator of progression of malignant melanoma. Carcinogenesis 32:1176–1182. https://doi.org/10.1093/carcin/bgr103
Ishiguro H, Uemura H, Fujinami K, Ikeda N, Ohta S, Kubota Y (2003) 55 kDa nuclear matrix protein (nmt55) mRNA is expressed in human prostate cancer tissue and is associated with the androgen receptor. Int J Cancer 105:26–32. https://doi.org/10.1002/ijc.11021
Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W (2013) CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res 41:e74. https://doi.org/10.1093/nar/gkt006
Yang P, Li J, Peng C et al (2020) TCONS_00012883 promotes proliferation and metastasis via DDX3/YY1/MMP1/PI3K-AKT axis in colorectal cancer. Clin Transl Med 10:e211. https://doi.org/10.1002/ctm2.211
Chen Z, Li JL, Lin S et al (2016) cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest 126:2267–2279. https://doi.org/10.1172/JCI85250
Chen Z, Lin S, Li JL et al (2018) CRTC1-MAML2 fusion-induced lncRNA LINC00473 expression maintains the growth and survival of human mucoepidermoid carcinoma cells. Oncogene 37:1885–1895. https://doi.org/10.1038/s41388-017-0104-0
Ma R, Zhai X, Zhu X, Zhang L (2019) LINC01585 functions as a regulator of gene expression by the CAMP/CREB signaling pathway in breast cancer. Gene 684:139–148. https://doi.org/10.1016/j.gene.2018.10.063
Qin Y, Chen W, Jiang G et al (2020) Interfering MSN-NONO complex-activated CREB signaling serves as a therapeutic strategy for triple-negative breast cancer. Sci Adv 6:eaaw9960. https://doi.org/10.1126/sciadv.aaw9960
Barba D, León-Sosa A, Lugo P et al (2021) Breast cancer, screening and diagnostic tools: all you need to know. Crit Rev Oncol Hematol 157:103174. https://doi.org/10.1016/j.critrevonc.2020.103174
Wang H, Li Y, Khan SA, Luo Y (2020) Prediction of breast cancer distant recurrence using natural language processing and knowledge-guided convolutional neural network. Artif Intell Med 110:101977. https://doi.org/10.1016/j.artmed.2020.101977
Peng JC, Shen J, Ran ZH (2013) Transcribed ultraconserved region in human cancers. RNA Biol 10:1771–1777. https://doi.org/10.4161/rna.26995
Chiang CW, Derti A, Schwartz D, Chou MF, Hirschhorn JN, Wu CT (2008) Ultraconserved elements: analyses of dosage sensitivity, motifs and boundaries. Genetics 180:2277–2293. https://doi.org/10.1534/genetics.108.096537
Calin GA, Liu CG, Ferracin M et al (2007) Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12:215–229. https://doi.org/10.1016/j.ccr.2007.07.027
Mestdagh P, Fredlund E, Pattyn F et al (2010) An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 29:3583–3592. https://doi.org/10.1038/onc.2010.106
Lujambio A, Portela A, Liz J et al (2010) CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 29:6390–6401. https://doi.org/10.1038/onc.2010.361
Ni W, Zhang Y, Zhan Z et al (2017) A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol 10:91. https://doi.org/10.1186/s13045-017-0449-4
Zhang LX, Xu L, Zhang CH, Lu YH, Ji TH, Ling LJ (2017) uc.38 induces breast cancer cell apoptosis via PBX1. Am J Cancer Res 7:2438–2451
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10:925–933. https://doi.org/10.4161/rna.24604
Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307. https://doi.org/10.1016/j.cell.2013.02.012
Guo CJ, Xu G, Chen LL (2020) Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci 45:947–960. https://doi.org/10.1016/j.tibs.2020.07.001
Turton N, Swan R, Mahenthiralingam T, Pitts D, Dykes IM (2019) The functions of Long Non-Coding RNA during embryonic cardiovascular development and its potential for diagnosis and treatment of congenital heart disease. J Cardiovasc Dev Dis. https://doi.org/10.3390/jcdd6020021
Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22:96–118. https://doi.org/10.1038/s41580-020-00315-9
Fatica A, Bozzoni I (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15:7–21. https://doi.org/10.1038/nrg3606
Wang X, Arai S, Song X et al (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454:126–130. https://doi.org/10.1038/nature06992
Deshar R, Yoo W, Cho EB, Kim S, Yoon JB (2019) RNF8 mediates NONO degradation following UV-induced DNA damage to properly terminate ATR-CHK1 checkpoint signaling. Nucleic Acids Res 47:762–778. https://doi.org/10.1093/nar/gky1166
Pavao M, Huang YH, Hafer LJ, Moreland RB, Traish AM (2001) Immunodetection of nmt55/p54nrb isoforms in human breast cancer. BMC Cancer 1:15. https://doi.org/10.1186/1471-2407-1-15
Traish AM, Huang YH, Ashba J et al (1997) Loss of expression of a 55-kDa nuclear protein (nmt55) in estrogen receptor-negative human breast cancer. Diagn Mol Pathol 6:209–221. https://doi.org/10.1097/00019606-199708000-00005
Iino K, Mitobe Y, Ikeda K et al (2020) RNA-binding protein NONO promotes breast cancer proliferation by post-transcriptional regulation of SKP2 and E2F8. Cancer Sci 111:148–159. https://doi.org/10.1111/cas.14240
Mayr B, Montminy M (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2:599–609. https://doi.org/10.1038/35085068
Wang H, Xu J, Lazarovici P, Quirion R, Zheng W (2018) cAMP response element-binding protein (CREB): a possible signaling molecule link in the pathophysiology of schizophrenia. Front Mol Neurosci 11:255. https://doi.org/10.3389/fnmol.2018.00255
Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855–859. https://doi.org/10.1038/365855a0
Cha-Molstad H, Keller DM, Yochum GS, Impey S, Goodman RH (2004) Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc Natl Acad Sci USA 101:13572–13577. https://doi.org/10.1073/pnas.0405587101
Johannessen M, Delghandi MP, Moens U (2004) What turns CREB on? Cell Signal 16:1211–1227. https://doi.org/10.1016/j.cellsig.2004.05.001
Xie F, Fan Q, Li BX, Xiao X (2019) Discovery of a synergistic inhibitor of cAMP-response element binding protein (CREB)-mediated gene transcription with 666–15. J Med Chem 62:11423–11429. https://doi.org/10.1021/acs.jmedchem.9b01207
Shankar DB, Cheng JC, Kinjo K et al (2005) The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 7:351–362. https://doi.org/10.1016/j.ccr.2005.02.018
Antonescu CR, Nafa K, Segal NH, Dal Cin P, Ladanyi M (2006) EWS-CREB1: a recurrent variant fusion in clear cell sarcoma–association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res 12:5356–5362. https://doi.org/10.1158/1078-0432.CCR-05-2811
Davis IJ, Kim JJ, Ozsolak F et al (2006) Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer Cell 9:473–484. https://doi.org/10.1016/j.ccr.2006.04.021
Amelio AL, Miraglia LJ, Conkright JJ et al (2007) A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci USA 104:20314–20319. https://doi.org/10.1073/pnas.0707999105
Acknowledgements
This research was supported in part by Jiangsu Province Six Talents Summit Project (WSW-001), Youth Talent Project (FRC201308), National Natural Science Foundation of China (81302305), and Natural Science Foundation of Jiangsu Province (BK20131027). We gratefully acknowledge the editorial assistance received from Changyan Ma, PhD.
Author information
Authors and Affiliations
Contributions
XS and XH contributed to the design and methodology of the study. XS and RC drafted the manuscript. YL, WZ, and YX participated in the acquisition and analysis of data. QZ was responsible for formal analysis. XZ and JW revised the study critically for important intellectual content and conducted quality control on it. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Ethics approval
All procedures were followed in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and those of the Helsinki Declaration. This work was approved by the ethics and research committee of the First Affiliated Hospital of Nanjing Medical University (2021-SRFA-175), and informed consent was obtained from all participants. Animal care and all the experimental protocols were approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University (IACUC-2002006–2).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, X., Huang, X., Chen, R. et al. The transcribed ultraconserved element uc.51 promotes the proliferation and metastasis of breast cancer by stabilizing NONO. Clin Exp Metastasis 38, 551–571 (2021). https://doi.org/10.1007/s10585-021-10128-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-021-10128-5