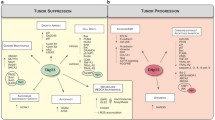
Abstract
Cancer largely adheres to Darwinian selection. Evolutionary forces are prominent during metastasis, the final and incurable disease stage, where cells acquire combinations of advantageous phenotypic features and interact with a dynamically changing microenvironment, in order to overcome the metastatic bottlenecks, while therapy exerts additional selective pressures. As a strategy to increase their fitness, tumors often co-opt developmental and tissue-homeostasis programs. Herein, 25 years after its discovery, we review TP73, a sibling of the cardinal tumor-suppressor TP53, through the lens of cancer evolution. The TP73 gene regulates a wide range of processes in embryonic development, tissue homeostasis and cancer via an overwhelming number of functionally divergent isoforms. We suggest that TP73 neither merely mimics TP53 via its p53-like tumor-suppressive functions, nor has black-or-white-type effects, as inferred by the antagonism between several of its isoforms in processes like apoptosis and DNA damage response. Rather, under dynamic conditions of selective pressure, the various p73 isoforms which are often co-expressed within the same cancer cells may work towards a common goal by simultaneously activating isoform-specific transcriptional and non-transcriptional programs. Combinatorial co-option of these programs offers selective advantages that overall increase the likelihood for successfully surpassing the barriers of the metastatic cascade. The p73 functional pleiotropy-based capabilities might be present in subclonal populations and expressed dynamically under changing microenvironmental conditions, thereby supporting clonal expansion and propelling evolution of metastasis. Deciphering the critical p73 isoform patterns along the spatiotemporal axes of tumor evolution could identify strategies to target TP73 for prevention and therapy of cancer metastasis.





Similar content being viewed by others
References
Yang, A., & McKeon, F. (2000). P63 and P73: P53 mimics, menaces and more. Nature Reviews Molecular Cell Biology, 1(3), 199–207. https://doi.org/10.1038/35043127
Graziano, V., & De Laurenzi, V. (2011). Role of p63 in cancer development. Biochimica et Biophysica Acta, 1816(1), 57–66. https://doi.org/10.1016/j.bbcan.2011.04.002
Su, X., Chakravarti, D., & Flores, E. R. (2013). p63 steps into the limelight: Crucial roles in the suppression of tumorigenesis and metastasis. Nature Reviews Cancer, 13(2), 136–143. https://doi.org/10.1038/nrc3446
Stiewe, T. (2007). The p53 family in differentiation and tumorigenesis. Nature Reviews Cancer, 7(3), 165–168. https://doi.org/10.1038/nrc2072
Li, Y., & Prives, C. (2007). Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene, 26(15), 2220–2225. https://doi.org/10.1038/sj.onc.1210311
Ramos, H., Raimundo, L., & Saraiva, L. (2020). p73: From the p53 shadow to a major pharmacological target in anticancer therapy. Pharmacological Research, 162, 105245. https://doi.org/10.1016/j.phrs.2020.105245
Stiewe, T., Theseling, C. C., & Pützer, B. M. (2002). Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: Implications for tumorigenesis. Journal of Biological Chemistry, 277(16), 14177–14185. https://doi.org/10.1074/jbc.M200480200
Kartasheva, N. N., Contente, A., Lenz-Stöppler, C., Roth, J., & Dobbelstein, M. (2002). p53 induces the expression of its antagonist p73 Delta N, establishing an autoregulatory feedback loop. Oncogene, 21(31), 4715–4727. https://doi.org/10.1038/sj.onc.1205584
Zaika, A. I., Slade, N., Erster, S. H., Sansome, C., Joseph, T. W., Pearl, M., et al. (2002). DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. Journal of Experimental Medicine, 196(6), 765–780. https://doi.org/10.1084/jem.20020179
Grob, T. J., Novak, U., Maisse, C., Barcaroli, D., Lüthi, A. U., Pirnia, F., et al. (2001). Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death and Differentiation, 8(12), 1213–1223. https://doi.org/10.1038/sj.cdd.4400962
Marabese, M., Vikhanskaya, F., & Broggini, M. (2007). p73: A chiaroscuro gene in cancer. European Journal of Cancer, 43(9), 1361–1372. https://doi.org/10.1016/j.ejca.2007.01.042
Tomasini, R., Tsuchihara, K., Wilhelm, M., Fujitani, M., Rufini, A., Cheung, C. C., et al. (2008). TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes and Development, 22(19), 2677–2691. https://doi.org/10.1101/gad.1695308
Wilhelm, M. T., Rufini, A., Wetzel, M. K., Tsuchihara, K., Inoue, S., Tomasini, R., et al. (2010). Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes and Development, 24(6), 549–560. https://doi.org/10.1101/gad.1873910
Stiewe, T., Zimmermann, S., Frilling, A., Esche, H., & Pützer, B. M. (2002). Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Research, 62(13), 3598–3602.
Steder, M., Alla, V., Meier, C., Spitschak, A., Pahnke, J., Fürst, K., et al. (2013). DNp73 exerts function in metastasis initiation by disconnecting the inhibitory role of EPLIN on IGF1R-AKT/STAT3 signaling. Cancer Cell, 24(4), 512–527. https://doi.org/10.1016/j.ccr.2013.08.023
Lunghi, P., Costanzo, A., Mazzera, L., Rizzoli, V., Levrero, M., & Bonati, A. (2009). The p53 family protein p73 provides new insights into cancer chemosensitivity and targeting. Clinical Cancer Research, 15(21), 6495–6502. https://doi.org/10.1158/1078-0432.CCR-09-1229
Amelio, I., Inoue, S., Markert, E. K., Levine, A. J., Knight, R. A., Mak, T. W., et al. (2015). TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1α degradation. Proc Natl Acad Sci U S A, 112(1), 226–231. https://doi.org/10.1073/pnas.1410609111
Stantic, M., Sakil, H. A., Zirath, H., Fang, T., Sanz, G., Fernandez-Woodbridge, A., et al. (2015). TAp73 suppresses tumor angiogenesis through repression of proangiogenic cytokines and HIF-1α activity. Proc Natl Acad Sci U S A, 112(1), 220–225. https://doi.org/10.1073/pnas.1421697112
Dulloo, I., Phang, B. H., Othman, R., Tan, S. Y., Vijayaraghavan, A., Goh, L. K., et al. (2015). Hypoxia-inducible TAp73 supports tumorigenesis by regulating the angiogenic transcriptome. Nature Cell Biology, 17(4), 511–523. https://doi.org/10.1038/ncb3130
López-Ferreras, L., Martínez-García, N., Maeso-Alonso, L., Martín-López, M., Díez-Matilla, Á., Villoch-Fernandez, J., et al. (2021). Deciphering the Nature of Trp73 Isoforms in Mouse Embryonic Stem Cell Models: Generation of Isoform-Specific. Cancers (Basel), 13, 13. https://doi.org/10.3390/cancers13133182
Gui, P., & Bivona, T. G. (2022). Evolution of metastasis: New tools and insights. Trends Cancer, 8(2), 98–109. https://doi.org/10.1016/j.trecan.2021.11.002
Merlo, L. M., Pepper, J. W., Reid, B. J., & Maley, C. C. (2006). Cancer as an evolutionary and ecological process. Nature Reviews Cancer, 6(12), 924–935. https://doi.org/10.1038/nrc2013
McGranahan, N., & Swanton, C. (2017). Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell, 168(4), 613–628. https://doi.org/10.1016/j.cell.2017.01.018
Birkbak, N. J., & McGranahan, N. (2020). Cancer genome evolutionary trajectories in metastasis. Cancer Cell, 37(1), 8–19. https://doi.org/10.1016/j.ccell.2019.12.004
Turajlic, S., & Swanton, C. (2016). Metastasis as an evolutionary process. Science, 352(6282), 169–175. https://doi.org/10.1126/science.aaf2784
Amirouchene-Angelozzi, N., Swanton, C., & Bardelli, A. (2017). Tumor evolution as a therapeutic target. Cancer Discov, https://doi.org/10.1158/2159-8290.Cd-17-0343
Rodrigues, P., Patel, S. A., Harewood, L., Olan, I., Vojtasova, E., Syafruddin, S. E., et al. (2018). NF-κB-dependent lymphoid enhancer co-option promotes renal carcinoma metastasis. Cancer Discovery, 8(7), 850–865. https://doi.org/10.1158/2159-8290.Cd-17-1211
Logotheti, S., Marquardt, S., Richter, C., Sophie Hain, R., Murr, N., Takan, I., et al. (2020). Neural networks recapitulation by cancer cells promotes disease progression: a novel role of p73 isoforms in cancer-neuronal crosstalk. Cancers, 12, 12. https://doi.org/10.3390/cancers12123789
Patel, S. A., Rodrigues, P., Wesolowski, L., & Vanharanta, S. (2021). Genomic control of metastasis. British Journal of Cancer, 124(1), 3–12. https://doi.org/10.1038/s41416-020-01127-6
Kerosuo, L., & Bronner-Fraser, M. (2012). What is bad in cancer is good in the embryo: Importance of EMT in neural crest development. Seminars in Cell & Developmental Biology, 23(3), 320–332. https://doi.org/10.1016/j.semcdb.2012.03.010
Rousseaux, S., Debernardi, A., Jacquiau, B., Vitte, A. L., Vesin, A., Nagy-Mignotte, H., et al. (2013). Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med, 5(186), 186ra166. https://doi.org/10.1126/scitranslmed.3005723
Richter, C., Marquardt, S., Li, F., Spitschak, A., Murr, N., Edelhäuser, B. A. H., et al. (2019). Rewiring E2F1 with classical NHEJ via APLF suppression promotes bladder cancer invasiveness. Journal of Experimental & Clinical Cancer Research, 38(1), 292. https://doi.org/10.1186/s13046-019-1286-9
Costanzo, V., Bardelli, A., Siena, S., & Abrignani, S. (2018). Exploring the links between cancer and placenta development. Open Biol, 8, 6. https://doi.org/10.1098/rsob.180081
Marquardt, S., Pavlopoulou, A., Takan, I., Dhar, P., Pützer, B. M., & Logotheti, S. (2021). A systems-based key innovation-driven approach infers co-option of jaw developmental programs during cancer progression. Front Cell Dev Biol, 9, 682619. https://doi.org/10.3389/fcell.2021.682619
Yılmaz, H., Toy, H. I., Marquardt, S., Karakülah, G., Küçük, C., Kontou, P. I., et al. (2021). In silico methods for the identification of diagnostic and favorable prognostic markers in acute myeloid leukemia. International Journal of Molecular Sciences, 22, 17. https://doi.org/10.3390/ijms22179601
Kerbel, R. S. (2000). Tumor angiogenesis: Past, present and the near future. Carcinogenesis, 21(3), 505–515. https://doi.org/10.1093/carcin/21.3.505
Cervantes-Villagrana, R. D., Albores-García, D., Cervantes-Villagrana, A. R., & García-Acevez, S. J. (2020). Tumor-induced neurogenesis and immune evasion as targets of innovative anti-cancer therapies. Signal Transduction and Targeted Therapy, 5(1), 99. https://doi.org/10.1038/s41392-020-0205-z
Mravec, B. (2022). Neurobiology of cancer: Definition, historical overview, and clinical implications. Cancer Medicine, 11(4), 903–921. https://doi.org/10.1002/cam4.4488
Martik, M. L., & Bronner, M. E. (2017). Regulatory logic underlying diversification of the neural crest. Trends in Genetics, 33(10), 715–727. https://doi.org/10.1016/j.tig.2017.07.015
Logotheti, S., & Pützer, B. M. (2019). STAT3 and STAT5 targeting for simultaneous management of melanoma and autoimmune diseases. Cancers (Basel), 11, 10. https://doi.org/10.3390/cancers11101448
Logotheti, S., Pavlopoulou, A., Galtsidis, S., Vojtesek, B., & Zoumpourlis, V. (2013). Functions, divergence and clinical value of TAp73 isoforms in cancer. Cancer and Metastasis Reviews, 32(3–4), 511–534. https://doi.org/10.1007/s10555-013-9424-x
Liu, G., Nozell, S., Xiao, H., & Chen, X. (2004). DeltaNp73beta is active in transactivation and growth suppression. Molecular and Cellular Biology, 24(2), 487–501. https://doi.org/10.1128/MCB.24.2.487-501.2004
Sakil, H. A. M., Stantic, M., Wolfsberger, J., Brage, S. E., Hansson, J., & Wilhelm, M. T. (2017). ΔNp73 regulates the expression of the multidrug-resistance genes ABCB1 and ABCB5 in breast cancer and melanoma cells - a short report. Cell Oncol (Dordr).https://doi.org/10.1007/s13402-017-0340-x
George, J., Lim, J. S., Jang, S. J., Cun, Y., Ozretić, L., Kong, G., et al. (2015). Comprehensive genomic profiles of small cell lung cancer. Nature, 524(7563), 47–53. https://doi.org/10.1038/nature14664
Stiewe, T., Tuve, S., Peter, M., Tannapfel, A., Elmaagacli, A. H., & Pützer, B. M. (2004). Quantitative TP73 transcript analysis in hepatocellular carcinomas. Clinical Cancer Research, 10(2), 626–633. https://doi.org/10.1158/1078-0432.ccr-0153-03
Osterburg, C., & Dötsch, V. (2022). Structural diversity of p63 and p73 isoforms. Cell Death and Differentiation. https://doi.org/10.1038/s41418-022-00975-4
Logotheti, S., Richter, C., Murr, N., Spitschak, A., Marquardt, S., & Putzer, B. M. (2021). Mechanisms of functional pleiotropy of p73 in cancer and beyond. Front Cell Dev Biol, 9, 737735. https://doi.org/10.3389/fcell.2021.737735
Koeppel, M., van Heeringen, S. J., Kramer, D., Smeenk, L., Janssen-Megens, E., Hartmann, M., et al. (2011). Crosstalk between c-Jun and TAp73alpha/beta contributes to the apoptosis-survival balance. Nucleic Acids Research, 39(14), 6069–6085. https://doi.org/10.1093/nar/gkr028
Oswald, C., & Stiewe, T. (2008). In good times and bad: P73 in cancer. Cell Cycle, 7(12), 1726–1731. https://doi.org/10.4161/cc.7.12.6148
Muppani, N., Nyman, U., & Joseph, B. (2011). TAp73alpha protects small cell lung carcinoma cells from caspase-2 induced mitochondrial mediated apoptotic cell death. Oncotarget, 2(12), 1145–1154. https://doi.org/10.18632/oncotarget.391
Cheng, C., Feng, S., Jiao, J., Huang, W., Huang, J., Wang, L., et al. (2018). DLC2 inhibits development of glioma through regulating the expression ratio of TAp73α/TAp73β. American Journal of Cancer Research, 8(7), 1200–1213.
Jiang, P., Du, W., & Yang, X. (2013). A critical role of glucose-6-phosphate dehydrogenase in TAp73-mediated cell proliferation. Cell Cycle, 12(24), 3720–3726. https://doi.org/10.4161/cc.27267
Velletri, T., Romeo, F., Tucci, P., Peschiaroli, A., Annicchiarico-Petruzzelli, M., Niklison-Chirou, M. V., et al. (2013). GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle, 12(22), 3564–3573. https://doi.org/10.4161/cc.26771
Amelio, I., Markert, E. K., Rufini, A., Antonov, A. V., Sayan, B. S., Tucci, P., et al. (2014). p73 regulates serine biosynthesis in cancer. Oncogene, 33(42), 5039–5046. https://doi.org/10.1038/onc.2013.456
Subramanian, D., Bunjobpol, W., & Sabapathy, K. (2015). Interplay between TAp73 protein and selected activator protein-1 (AP-1) family members promotes AP-1 target gene activation and cellular Growth. Journal of Biological Chemistry, 290(30), 18636–18649. https://doi.org/10.1074/jbc.M115.636548
Nemajerova, A., & Moll, U. M. (2019). Tissue-specific roles of p73 in development and homeostasis. Journal of Cell Science, 132, 19. https://doi.org/10.1242/jcs.233338
Fernandez-Alonso, R., Martin-Lopez, M., Gonzalez-Cano, L., Garcia, S., Castrillo, F., Diez-Prieto, I., et al. (2015). p73 is required for endothelial cell differentiation, migration and the formation of vascular networks regulating VEGF and TGFβ signaling. Cell Death and Differentiation, 22(8), 1287–1299. https://doi.org/10.1038/cdd.2014.214
Sabapathy, K. (2015). p73: A positive or negative regulator of angiogenesis, or both? Molecular and Cellular Biology, 36(6), 848–854. https://doi.org/10.1128/MCB.00929-15
Dulloo, I., Hooi, P. B., & Sabapathy, K. (2015). Hypoxia-induced DNp73 stabilization regulates Vegf-A expression and tumor angiogenesis similar to TAp73. Cell Cycle, 14(22), 3533–3539. https://doi.org/10.1080/15384101.2015.1078038
He, Z., Agostini, M., Liu, H., Melino, G., & Simon, H. U. (2015). p73 regulates basal and starvation-induced liver metabolism in vivo. Oncotarget, 6(32), 33178–33190. https://doi.org/10.18632/oncotarget.5090
Amelio, I., Antonov, A. A., Catani, M. V., Massoud, R., Bernassola, F., Knight, R. A., et al. (2014). TAp73 promotes anabolism. Oncotarget, 5(24), 12820–12934. https://doi.org/10.18632/oncotarget.2667
Yang, A., Walker, N., Bronson, R., Kaghad, M., Oosterwegel, M., Bonnin, J., et al. (2000). p73-Deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature, 404(6773), 99–103. https://doi.org/10.1038/35003607
Tomasini, R., Secq, V., Pouyet, L., Thakur, A. K., Wilhelm, M., Nigri, J., et al. (2013). TAp73 is required for macrophage-mediated innate immunity and the resolution of inflammatory responses. Cell Death and Differentiation, 20(2), 293–301. https://doi.org/10.1038/cdd.2012.123
Koshiba, S., Ichimiya, S., Nagashima, T., Tonooka, A., Kubo, T., Kikuchi, T., et al. (2008). Tonsillar crypt epithelium of palmoplantar pustulosis secretes interleukin-6 to support B-cell development via p63/p73 transcription factors. The Journal of Pathology, 214(1), 75–84. https://doi.org/10.1002/path.2266
Kumagai, A., Kubo, T., Kawata, K., Kamekura, R., Yamashita, K., Jitsukawa, S., et al. (2017). Keratinocytes in atopic dermatitis express abundant ΔNp73 regulating thymic stromal lymphopoietin production via NF-κB. Journal of Dermatological Science, 88(2), 175–183. https://doi.org/10.1016/j.jdermsci.2017.06.017
Vikhreva, P., Petrova, V., Gokbulut, T., Pestlikis, I., Mancini, M., Di Daniele, N., et al. (2017). TAp73 upregulates IL-1β in cancer cells: Potential biomarker in lung and breast cancer? Biochemical and Biophysical Research Communications, 482(3), 498–505. https://doi.org/10.1016/j.bbrc.2016.10.085
Bent, R., Moll, L., Grabbe, S., & Bros, M. (2018). Interleukin-1 Beta-A friend or foe in malignancies? International Journal of Molecular Sciences, 19, 8. https://doi.org/10.3390/ijms19082155
Wolfsberger, J., Sakil, H. A. M., Zhou, L., van Bree, N., Baldisseri, E., de Souza Ferreira, S., et al. (2021). TAp73 represses NF-κB-mediated recruitment of tumor-associated macrophages in breast cancer. Proceedings of the National Academy of Sciences, 118, 10. https://doi.org/10.1073/pnas.2017089118
Rozenberg, J. M., Zvereva, S., Dalina, A., Blatov, I., Zubarev, I., Luppov, D., et al. (2021). Dual role of p73 in cancer microenvironment and dna damage response. Cells, 10, 12. https://doi.org/10.3390/cells10123516
Ren, M., Kazemian, M., Zheng, M., He, J., Li, P., Oh, J., et al. (2020). Transcription factor p73 regulates Th1 differentiation. Nature Communications, 11(1), 1475. https://doi.org/10.1038/s41467-020-15172-5
Niklison-Chirou, M. V., Agostini, M., Amelio, I., & Melino, G. (2020). Regulation of adult neurogenesis in mammalian brain. International Journal of Molecular Sciences, 21, 14. https://doi.org/10.3390/ijms21144869
Griffin, N., Faulkner, S., Jobling, P., & Hondermarck, H. (2018). Targeting neurotrophin signaling in cancer: The renaissance. Pharmacological Research, 135, 12–17. https://doi.org/10.1016/j.phrs.2018.07.019
Monje, M., Borniger, J. C., D’Silva, N. J., Deneen, B., Dirks, P. B., Fattahi, F., et al. (2020). Roadmap for the emerging field of cancer neuroscience. Cell, 181(2), 219–222. https://doi.org/10.1016/j.cell.2020.03.034
Friedmann-Morvinski, D., & Verma, I. M. (2014). Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Reports, 15(3), 244–253. https://doi.org/10.1002/embr.201338254
Batlle, E., & Clevers, H. (2017). Cancer stem cells revisited. Nature Medicine, 23(10), 1124–1134. https://doi.org/10.1038/nm.4409
Talos, F., Abraham, A., Vaseva, A. V., Holembowski, L., Tsirka, S. E., Scheel, A., et al. (2010). p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death and Differentiation, 17(12), 1816–1829. https://doi.org/10.1038/cdd.2010.131
Fujitani, M., Cancino, G. I., Dugani, C. B., Weaver, I. C., Gauthier-Fisher, A., Paquin, A., et al. (2010). TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Current Biology, 20(22), 2058–2065. https://doi.org/10.1016/j.cub.2010.10.029
Gonzalez-Cano, L., Herreros-Villanueva, M., Fernandez-Alonso, R., Ayuso-Sacido, A., Meyer, G., Garcia-Verdugo, J. M., et al. (2010). p73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death & Disease, 1, e109. https://doi.org/10.1038/cddis.2010.87
Agostini, M., Tucci, P., Chen, H., Knight, R. A., Bano, D., Nicotera, P., et al. (2010). p73 regulates maintenance of neural stem cell. Biochemical and Biophysical Research Communications, 403(1), 13–17. https://doi.org/10.1016/j.bbrc.2010.10.087
Killick, R., Niklison-Chirou, M., Tomasini, R., Bano, D., Rufini, A., Grespi, F., et al. (2011). p73: A multifunctional protein in neurobiology. Molecular Neurobiology, 43(2), 139–146. https://doi.org/10.1007/s12035-011-8172-6
Meier, C., Hardtstock, P., Joost, S., Alla, V., & Pützer, B. M. (2016). p73 and IGF1R regulate emergence of aggressive cancer stem-like features via miR-885-5p control. Cancer Research, 76(2), 197–205. https://doi.org/10.1158/0008-5472.CAN-15-1228
Galtsidis, S., Logotheti, S., Pavlopoulou, A., Zampetidis, C. P., Papachristopoulou, G., Scorilas, A., et al. (2017). Unravelling a p73-regulated network: The role of a novel p73-dependent target, MIR3158, in cancer cell migration and invasiveness. Cancer Letters, 388, 96–106. https://doi.org/10.1016/j.canlet.2016.11.036
Daskalos, A., Logotheti, S., Markopoulou, S., Xinarianos, G., Gosney, J. R., Kastania, A. N., et al. (2011). Global DNA hypomethylation-induced ΔNp73 transcriptional activation in non-small cell lung cancer. Cancer Letters, 300(1), 79–86. https://doi.org/10.1016/j.canlet.2010.09.009
Logotheti, S., Michalopoulos, I., Sideridou, M., Daskalos, A., Kossida, S., Spandidos, D. A., et al. (2010). Sp1 binds to the external promoter of the p73 gene and induces the expression of TAp73gamma in lung cancer. FEBS Journal, 277(14), 3014–3027. https://doi.org/10.1111/j.1742-4658.2010.07710.x
Fürst, K., Steder, M., Logotheti, S., Angerilli, A., Spitschak, A., Marquardt, S., et al. (2019). DNp73-induced degradation of tyrosinase links depigmentation with EMT-driven melanoma progression. Cancer Letters, 442, 299–309. https://doi.org/10.1016/j.canlet.2018.11.009
Sayan, A. E., Sayan, B. S., Findikli, N., & Ozturk, M. (2001). Acquired expression of transcriptionally active p73 in hepatocellular carcinoma cells. Oncogene, 20(37), 5111–5117. https://doi.org/10.1038/sj.onc.1204669
Woodstock, D. L., Sammons, M. A., & Fischer, M. (2021). p63 and p53: Collaborative partners or dueling rivals? Front Cell Dev Biol, 9, 701986. https://doi.org/10.3389/fcell.2021.701986
Coutandin, D., Löhr, F., Niesen, F. H., Ikeya, T., Weber, T. A., Schäfer, B., et al. (2009). Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death and Differentiation, 16(12), 1582–1589. https://doi.org/10.1038/cdd.2009.139
Joerger, A. C., Rajagopalan, S., Natan, E., Veprintsev, D. B., Robinson, C. V., & Fersht, A. R. (2009). Structural evolution of p53, p63, and p73: Implication for heterotetramer formation. Proc Natl Acad Sci U S A, 106(42), 17705–17710. https://doi.org/10.1073/pnas.0905867106
Gebel, J., Luh, L. M., Coutandin, D., Osterburg, C., Löhr, F., Schäfer, B., et al. (2016). Mechanism of TAp73 inhibition by ΔNp63 and structural basis of p63/p73 hetero-tetramerization. Cell Death and Differentiation, 23(12), 1930–1940. https://doi.org/10.1038/cdd.2016.83
Rocco, J. W., Leong, C. O., Kuperwasser, N., DeYoung, M. P., & Ellisen, L. W. (2006). p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell, 9(1), 45–56. https://doi.org/10.1016/j.ccr.2005.12.013
Marin, M. C., Jost, C. A., Brooks, L. A., Irwin, M. S., O’Nions, J., Tidy, J. A., et al. (2000). A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nature Genetics, 25(1), 47–54. https://doi.org/10.1038/75586
Gaiddon, C., Lokshin, M., Ahn, J., Zhang, T., & Prives, C. (2001). A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Molecular and Cellular Biology, 21(5), 1874–1887. https://doi.org/10.1128/MCB.21.5.1874-1887.2001
Stindt, M. H., Muller, P. A., Ludwig, R. L., Kehrloesser, S., Dötsch, V., & Vousden, K. H. (2015). Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene, 34(33), 4300–4310. https://doi.org/10.1038/onc.2014.359
Kehrloesser, S., Osterburg, C., Tuppi, M., Schäfer, B., Vousden, K. H., & Dötsch, V. (2016). Intrinsic aggregation propensity of the p63 and p73 TI domains correlates with p53R175H interaction and suggests further significance of aggregation events in the p53 family. Cell Death and Differentiation, 23(12), 1952–1960. https://doi.org/10.1038/cdd.2016.75
Xu, J., Reumers, J., Couceiro, J. R., De Smet, F., Gallardo, R., Rudyak, S., et al. (2011). Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nature Chemical Biology, 7(5), 285–295. https://doi.org/10.1038/nchembio.546
Petronilho, E. C., Pedrote, M. M., Marques, M. A., Passos, Y. M., Mota, M. F., Jakobus, B., et al. (2021). Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands. Chemical Science, 12(21), 7334–7349. https://doi.org/10.1039/d1sc01739j
Wang, G., & Fersht, A. R. (2017). Multisite aggregation of p53 and implications for drug rescue. Proc Natl Acad Sci U S A, 114(13), E2634–E2643. https://doi.org/10.1073/pnas.1700308114
Anbarasan, T., & Bourdon, J. C. (2019). The emerging landscape of p53 isoforms in physiology, cancer and degenerative diseases. International Journal of Molecular Sciences, 20, 24. https://doi.org/10.3390/ijms20246257
Zorić, A., Horvat, A., & Slade, N. (2013). Differential effects of diverse p53 isoforms on TAp73 transcriptional activity and apoptosis. Carcinogenesis, 34(3), 522–529. https://doi.org/10.1093/carcin/bgs370
Zhang, J., Sun, W., Kong, X., Zhang, Y., Yang, H. J., Ren, C., et al. (2019). Mutant p53 antagonizes p63/p73-mediated tumor suppression via Notch1. Proceedings of the National Academy of Sciences of the United States of America, 116(48), 24259–24267. https://doi.org/10.1073/pnas.1913919116
Slade, N., Zaika, A. I., Erster, S., & Moll, U. M. (2004). DeltaNp73 stabilises TAp73 proteins but compromises their function due to inhibitory hetero-oligomer formation. Cell Death and Differentiation, 11(3), 357–360. https://doi.org/10.1038/sj.cdd.4401335
Ferraiuolo, M., Di Agostino, S., Blandino, G., & Strano, S. (2016). Oncogenic intra-p53 family member interactions in human cancers. Frontiers in Oncology, 6, 77. https://doi.org/10.3389/fonc.2016.00077
Nemajerova, A., Amelio, I., Gebel, J., Dötsch, V., Melino, G., & Moll, U. M. (2018). Non-oncogenic roles of TAp73: From multiciliogenesis to metabolism. Cell Death and Differentiation, 25(1), 144–153. https://doi.org/10.1038/cdd.2017.178
Tang, Q., Su, Z., Gu, W., & Rustgi, A. K. (2020). Mutant p53 on the path to metastasis. Trends Cancer, 6(1), 62–73. https://doi.org/10.1016/j.trecan.2019.11.004
Carroll, D. K., Carroll, J. S., Leong, C. O., Cheng, F., Brown, M., Mills, A. A., et al. (2006). p63 regulates an adhesion programme and cell survival in epithelial cells. Nature Cell Biology, 8(6), 551–561. https://doi.org/10.1038/ncb1420
Barbieri, C. E., Tang, L. J., Brown, K. A., & Pietenpol, J. A. (2006). Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Research, 66(15), 7589–7597. https://doi.org/10.1158/0008-5472.CAN-06-2020
Olive, K. P., Tuveson, D. A., Ruhe, Z. C., Yin, B., Willis, N. A., Bronson, R. T., et al. (2004). Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell, 119(6), 847–860. https://doi.org/10.1016/j.cell.2004.11.004
Lang, G. A., Iwakuma, T., Suh, Y. A., Liu, G., Rao, V. A., Parant, J. M., et al. (2004). Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell, 119(6), 861–872. https://doi.org/10.1016/j.cell.2004.11.006
Aubrey, B. J., Janic, A., Chen, Y., Chang, C., Lieschke, E. C., Diepstraten, S. T., et al. (2018). Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes and Development, 32(21–22), 1420–1429. https://doi.org/10.1101/gad.314286.118
Amelio, I., Panatta, E., Niklison-Chirou, M. V., Steinert, J. R., Agostini, M., Morone, N., et al. (2020). The C terminus of p73 is essential for hippocampal development. Proc Natl Acad Sci U S A, 117(27), 15694–15701. https://doi.org/10.1073/pnas.2000917117
Laubach, K. N., Yan, W., Kong, X., Sun, W., Chen, M., Zhang, J., et al. (2022). p73α1, a p73 C-terminal isoform, regulates tumor suppression and the inflammatory response via Notch1. Proc Natl Acad Sci U S A, 119(22), e2123202119. https://doi.org/10.1073/pnas.2123202119
Denny, S. K., Yang, D., Chuang, C. H., Brady, J. J., Lim, J. S., Grüner, B. M., et al. (2016). Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell, 166(2), 328–342. https://doi.org/10.1016/j.cell.2016.05.052
Baccin, C., Al-Sabah, J., Velten, L., Helbling, P. M., Grünschläger, F., Hernández-Malmierca, P., et al. (2020). Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nature Cell Biology, 22(1), 38–48. https://doi.org/10.1038/s41556-019-0439-6
Das, S., & Somasundaram, K. (2006). Therapeutic potential of an adenovirus expressing p73 beta, a p53 homologue, against human papilloma virus positive cervical cancer in vitro and in vivo. Cancer Biology & Therapy, 5(2), 210–217. https://doi.org/10.4161/cbt.5.2.2402
Andrews, M. C., & Wargo, J. A. (2017). Cancer evolution during immunotherapy. Cell, 171(4), 740–742. https://doi.org/10.1016/j.cell.2017.10.027
Zehir, A., Benayed, R., Shah, R. H., Syed, A., Middha, S., Kim, H. R., et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature Medicine, 23(6), 703–713. https://doi.org/10.1038/nm.4333
Ghatak, D., Das Ghosh, D., & Roychoudhury, S. (2020). Cancer stemness: P53 at the wheel. Frontiers in Oncology, 10, 604124. https://doi.org/10.3389/fonc.2020.604124
Liu, J., Zhang, C., Hu, W., & Feng, Z. (2015). Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Letters, 356(2 Pt A), 197–203. https://doi.org/10.1016/j.canlet.2013.12.025
Ghosh, M., Saha, S., Bettke, J., Nagar, R., Parrales, A., Iwakuma, T., et al. (2021). Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell, 39(4), 494-508.e495. https://doi.org/10.1016/j.ccell.2021.01.003
Cooks, T., Pateras, I. S., Tarcic, O., Solomon, H., Schetter, A. J., Wilder, S., et al. (2013). Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell, 23(5), 634–646. https://doi.org/10.1016/j.ccr.2013.03.022
Alvarado-Ortiz, E., de la Cruz-López, K. G., Becerril-Rico, J., Sarabia-Sánchez, M. A., Ortiz-Sánchez, E., & García-Carrancá, A. (2020). Mutant p53 gain-of-function: Role in cancer development, progression, and therapeutic approaches. Front Cell Dev Biol, 8, 607670. https://doi.org/10.3389/fcell.2020.607670
Moses, M. A., George, A. L., Sakakibara, N., Mahmood, K., Ponnamperuma, R. M., King, K. E., et al. (2019). Molecular mechanisms of p63-mediated squamous cancer pathogenesis. International Journal of Molecular Sciences, 20, 14. https://doi.org/10.3390/ijms20143590
Bid, H. K., Roberts, R. D., Cam, M., Audino, A., Kurmasheva, R. T., Lin, J., et al. (2014). ΔNp63 promotes pediatric neuroblastoma and osteosarcoma by regulating tumor angiogenesis. Cancer Research, 74(1), 320–329. https://doi.org/10.1158/0008-5472.CAN-13-0894
Gatti, V., Fierro, C., Annicchiarico-Petruzzelli, M., Melino, G., & Peschiaroli, A. (2019). ΔNp63 in squamous cell carcinoma: Defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Molecular Oncology, 13(5), 981–1001. https://doi.org/10.1002/1878-0261.12473
Acknowledgements
All figures presented in this work were created using BioRender (biorender.com). AGG acknowledges Deutscher Akademischer Austauschdienst “Hochschulpartnerschaften mit Griechenland” (No. 57513880) and “DNA Damage and Repair and Their Relevance to Carcinogenesis” (No. 57339330).
Funding
This work was supported by Deutsche Forschungsgemeinschaft, TRR81/3 109546710 Project A10 to TS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Logotheti, S., Pavlopoulou, A., Marquardt, S. et al. p73 isoforms meet evolution of metastasis. Cancer Metastasis Rev 41, 853–869 (2022). https://doi.org/10.1007/s10555-022-10057-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-022-10057-z