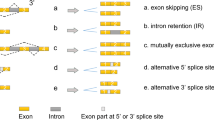
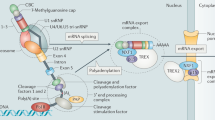
Abstract
The protein-coding regions of mRNAs have the information to make proteins and hence have been at the center of attention for understanding altered protein functions in disease states, including cancer. Indeed, the discovery of genomic alterations and driver mutations that change protein levels and/or activity has been pivotal in our understanding of cancer biology. However, to better understand complex molecular mechanisms that are deregulated in cancers, we also need to look at non-coding parts of mRNAs, including 3′UTRs (untranslated regions), which control mRNA stability, localization, and translation efficiency. Recently, these rather overlooked regions of mRNAs are gaining attention as mounting evidence provides functional links between 3′UTRs, protein functions, and cancer-related molecular mechanisms. Here, roles of 3′UTRs in cancer biology and mechanisms that result in cancer-specific 3′-end isoform variants will be reviewed. An increased appreciation of 3′UTRs may help the discovery of new ways to explain as of yet unknown oncogene activation and tumor suppressor inactivation cases in cancers, and provide new avenues for diagnostic and therapeutic applications.



Similar content being viewed by others
References
Nangalia, J., & Campbell, P. J. (2019). Genome sequencing during a patient’s journey through cancer. The New England Journal of Medicine, 381(22), 2145–2156. https://doi.org/10.1056/NEJMra1910138.
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science, 309(5740), 1559–1563. https://doi.org/10.1126/science.1112014.
Huang, Q., Yan, J., & Agami, R. (2018). Long non-coding RNAs in metastasis. Cancer Metastasis Reviews, 37(1), 75–81. https://doi.org/10.1007/s10555-017-9713-x.
Anastasiadou, E., Jacob, L. S., & Slack, F. J. (2018). Non-coding RNA networks in cancer. Nature Reviews. Cancer, 18(1), 5–18. https://doi.org/10.1038/nrc.2017.99.
Yang, E., van Nimwegen, E., Zavolan, M., Rajewsky, N., Schroeder, M., Magnasco, M., & Darnell JE Jr. (2003). Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Research, 13(8), 1863–1872. https://doi.org/10.1101/gr.1272403.
Sharova, L. V., Sharov, A. A., Nedorezov, T., Piao, Y., Shaik, N., & Ko, M. S. (2009). Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Research, 16(1), 45–58. https://doi.org/10.1093/dnares/dsn030.
Friedel, C. C., & Dölken, L. (2009). Metabolic tagging and purification of nascent RNA: Implications for transcriptomics. Molecular BioSystems, 5(11), 1271–1278. https://doi.org/10.1039/b911233b.
Eser, P., Demel, C., Maier, K. C., Schwalb, B., Pirkl, N., Martin, D. E., Cramer, P., & Tresch, A. (2014). Periodic mRNA synthesis and degradation co-operate during cell cycle gene expression. Molecular Systems Biology, 10, 717. https://doi.org/10.1002/msb.134886.
Gerstberger, S., Hafner, M., & Tuschl, T. (2014). A census of human RNA-binding proteins. Nature Reviews. Genetics, 15(12), 829–845. https://doi.org/10.1038/nrg3813.
Lunde, B. M., Moore, C., & Varani, G. (2007). RNA-binding proteins: Modular design for efficient function. Nature Reviews. Molecular Cell Biology, 8(6), 479–490. https://doi.org/10.1038/nrm2178.
Hentze, M. W., Castello, A., Schwarzl, T., & Preiss, T. (2018). A brave new world of RNA-binding proteins. Nature Reviews. Molecular Cell Biology, 19(5), 327–341. https://doi.org/10.1038/nrm.2017.130.
Lewis, B. P., Burge, C. B., & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120(1), 15–20. https://doi.org/10.1016/j.cell.2004.12.035.
Paulsen, M. T., Veloso, A., Prasad, J., Bedi, K., Ljungman, E. A., Tsan, Y. C., Chang, C. W., Tarrier, B., Washburn, J. G., Lyons, R., Robinson, D. R., Kumar-Sinha, C., Wilson, T. E., & Ljungman, M. (2013). Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proceedings of the National Academy of Sciences of the United States of America, 110(6), 2240–2245. https://doi.org/10.1073/pnas.1219192110.
Cannell, I. G., Merrick, K. A., Morandell, S., Zhu, C. Q., Braun, C. J., Grant, R. A., Cameron, E. R., Tsao, M. S., Hemann, M. T., & Yaffe, M. B. (2015). A pleiotropic RNA-binding protein controls distinct cell cycle checkpoints to drive resistance of p53-defective tumors to chemotherapy. Cancer Cell, 28, 623–637. https://doi.org/10.1016/j.ccell.2015.09.009.
Medford, R. M., Nguyen, H. T., & Nadal-Ginard, B. (1983). Transcriptional and cell cycle-mediated regulation of myosin heavy chain gene expression during muscle cell differentiation. The Journal of Biological Chemistry, 258(18), 11063–11073.
Hitti, E., Iakovleva, T., Brook, M., Deppenmeier, S., Gruber, A. D., Radzioch, D., Clark, A. R., Blackshear, P. J., Kotlyarov, A., & Gaestel, M. (2006). Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Molecular and Cellular Biology, 26(6), 2399–2407. https://doi.org/10.1128/MCB.26.6.2399-2407.2006.
Abdelmohsen, K., & Gorospe, M. (2010). Posttranscriptional regulation of cancer traits by HuR. Wiley Interdisciplinary Reviews RNA, 1(2), 214–229. https://doi.org/10.1002/wrna.4.
Sengupta, S., Jang, B. C., Wu, M. T., Paik, J. H., Furneaux, H., & Hla, T. (2003). The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. The Journal of Biological Chemistry, 278(27), 25227–25233. https://doi.org/10.1074/jbc.M301813200.
Torun, A., Enayat, S., Sheraj, I., Tunçer, S., Ülgen, D. H., & Banerjee, S. (2019). Butyrate mediated regulation of RNA binding proteins in the post-transcriptional regulation of inflammatory gene expression. Cellular Signalling, 64, 109410. https://doi.org/10.1016/j.cellsig.2019.109410.
Levine, A. J. (1997). p53, the cellular gatekeeper for growth and division. Cell, 88(3), 323–331. https://doi.org/10.1016/s0092-8674(00)81871-1.
Hollstein, M., Sidransky, D., Vogelstein, B., & Harris, C. C. (1991). p53 mutations in human cancers. Science, 253(5015), 49–53. https://doi.org/10.1126/science.1905840.
Soussi, T., & Wiman, K. G. (2007). Shaping genetic alterations in human cancer: The p53 mutation paradigm. Cancer Cell, 12(4), 303–312. https://doi.org/10.1016/j.ccr.2007.10.001.
Morandell, S., Reinhardt, H. C., Cannell, I. G., Kim, J. S., Ruf, D. M., Mitra, T., Couvillon, A. D., Jacks, T., & Yaffe, M. B. (2013). A reversible gene-targeting strategy identifies synthetic lethal interactions between MK2 and p53 in the DNA damage response in vivo. Cell Reports, 5(4), 868–877. https://doi.org/10.1016/j.celrep.2013.10.025.
Reinhardt, H. C., Hasskamp, P., Schmedding, I., Morandell, S., van Vugt, M. A., Wang, X., et al. (2010). DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Molecular Cell, 40(1), 34–49. https://doi.org/10.1016/j.molcel.2010.09.018.
Rosenstierne, M. W., Vinther, J., Mittler, G., Larsen, L., Mann, M., & Norrild, B. (2008). Conserved CPEs in the p53 3′ untranslated region influence mRNA stability and protein synthesis. Anticancer Research, 28(5A), 2553–2559.
Vilborg, A., Glahder, J. A., Wilhelm, M. T., Bersani, C., Corcoran, M., Mahmoudi, S., Rosenstierne, M., Grandér, D., Farnebo, M., Norrild, B., & Wiman, K. G. (2009). The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proceedings of the National Academy of Sciences of the United States of America, 106(37), 15756–15761. https://doi.org/10.1073/pnas.0900862106.
Mazan-Mamczarz, K., Galbán, S., López de Silanes, I., Martindale, J. L., Atasoy, U., Keene, J. D., et al. (2003). RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proceedings of the National Academy of Sciences of the United States of America, 100(14), 8354–8359. https://doi.org/10.1073/pnas.1432104100.
Ahuja, D., Goyal, A., & Ray, P. S. (2016). Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biology, 13(11), 1152–1165. https://doi.org/10.1080/15476286.2016.1229734.
Liu, L., Ouyang, M., Rao, J. N., Zou, T., Xiao, L., Chung, H. K., Wu, J., Donahue, J. M., Gorospe, M., & Wang, J. Y. (2015). Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Molecular Biology of the Cell, 26(10), 1797–1810. https://doi.org/10.1091/mbc.E14-11-1500.
Mukherjee, N., Jacobs, N. C., Hafner, M., Kennington, E. A., Nusbaum, J. D., Tuschl, T., Blackshear, P. J., & Ohler, U. (2014). Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biology, 15(1), R12. https://doi.org/10.1186/gb-2014-15-1-r12.
Kuwano, Y., Nishida, K., Kajita, K., Satake, Y., Akaike, Y., Fujita, K., Kano, S., Masuda, K., & Rokutan, K. (2015). Transformer 2β and miR-204 regulate apoptosis through competitive binding to 3′ UTR of BCL2 mRNA. Cell Death and Differentiation, 22(5), 815–825. https://doi.org/10.1038/cdd.2014.176.
Hennig, J., & Sattler, M. (2015). Deciphering the protein-RNA recognition code: Combining large-scale quantitative methods with structural biology. Bioessays, 37(8), 899–908. https://doi.org/10.1002/bies.201500033.
Cho, S. J., Zhang, J., & Chen, X. (2010). RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Research, 38(7), 2256–2267. https://doi.org/10.1093/nar/gkp1229.
Torres-Fernández, L. A., Jux, B., Bille, M., Port, Y., Schneider, K., Geyer, M., Mayer, G., & Kolanus, W. (2019). The mRNA repressor TRIM71 cooperates with nonsense-mediated decay factors to destabilize the mRNA of CDKN1A/p21. Nucleic Acids Research, 47(22), 11861–11879. https://doi.org/10.1093/nar/gkz1057.
Kurosaki, T., Popp, M. W., & Maquat, L. E. (2019). Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nature Reviews. Molecular Cell Biology, 20(7), 406–420. https://doi.org/10.1038/s41580-019-0126-2.
Park, J., Seo, J. W., Ahn, N., Park, S., Hwang, J., & Nam, J. W. (2019). UPF1/SMG7-dependent microRNA-mediated gene regulation. Nature Communications, 10(1), 4181. https://doi.org/10.1038/s41467-019-12123-7.
Kim, Y. K., & Maquat, L. E. (2019). UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA, 25(4), 407–422. https://doi.org/10.1261/rna.070136.118.
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297.
Erson, A. E., & Petty, E. M. (2008). MicroRNAs in development and disease. Clinical Genetics, 74(4), 296–306. https://doi.org/10.1111/j.1399-0004.2008.01076.x.
Tuna, M., Machado, A. S., & Calin, G. A. (2016). Genetic and epigenetic alterations of microRNAs and implications for human cancers and other diseases. Genes, Chromosomes & Cancer, 55(3), 193–214. https://doi.org/10.1002/gcc.22332.
Gebert, L. F. R., & MacRae, I. J. (2019). Regulation of microRNA function in animals. Nature Reviews. Molecular Cell Biology, 20(1), 21–37. https://doi.org/10.1038/s41580-018-0045-7.
Peng, Y., & Croce, C. M. (2016). The role of MicroRNAs in human cancer. Signal Transduction and Targeted Therapy, 1, 15004. https://doi.org/10.1038/sigtrans.2015.4.
Teige, I., Mårtensson, L., & Frendéus, B. L. (2019). Targeting the antibody checkpoints to enhance cancer immunotherapy-focus on FcγRIIB. Frontiers in Immunology, 10, 481. https://doi.org/10.3389/fimmu.2019.00481.
Hermeking, H. (2007). p53 enters the microRNA world. Cancer Cell, 12(5), 414–418.
Zhang, L., Liao, Y., & Tang, L. (2019). MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. Journal of Experimental & Clinical Cancer Research, 38(1), 53. https://doi.org/10.1186/s13046-019-1059-5.
Slabáková, E., Culig, Z., Remšík, J., & Souček, K. (2017). Alternative mechanisms of miR-34a regulation in cancer. Cell Death & Disease, 8(10), e3100. https://doi.org/10.1038/cddis.2017.495.
Chen, P. C., Yu, C. C., Huang, W. Y., Huang, W. H., Chuang, Y. M., Lin, R. I., et al. (2019). c-MYC acts as a competing endogenous RNA to sponge. Cancers (Basel), 11(10). https://doi.org/10.3390/cancers11101457.
Szostak, E., & Gebauer, F. (2013). Translational control by 3'-UTR-binding proteins. Briefings in Functional Genomics, 12(1), 58–65. https://doi.org/10.1093/bfgp/els056.
Uniacke, J., Holterman, C. E., Lachance, G., Franovic, A., Jacob, M. D., Fabian, M. R., Payette, J., Holcik, M., Pause, A., & Lee, S. (2012). An oxygen-regulated switch in the protein synthesis machinery. Nature, 486(7401), 126–129. https://doi.org/10.1038/nature11055.
Szostak, E., García-Beyaert, M., Guitart, T., Graindorge, A., Coll, O., & Gebauer, F. (2018). Hrp48 and eIF3d contribute to msl-2 mRNA translational repression. Nucleic Acids Research, 46(8), 4099–4113. https://doi.org/10.1093/nar/gky246.
Arake de Tacca, L. M., Pulos-Holmes, M. C., Floor, S. N., & Cate, J. H. D. (2019). PTBP1 mRNA isoforms and regulation of their translation. RNA, 25(10), 1324–1336. https://doi.org/10.1261/rna.070193.118.
Mazumder, B., Sampath, P., & Fox, P. L. (2005). Regulation of macrophage ceruloplasmin gene expression: One paradigm of 3'-UTR-mediated translational control. Molecules and Cells, 20(2), 167–172.
Ray, P. S., & Fox, P. L. (2007). A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. The EMBO Journal, 26(14), 3360–3372. https://doi.org/10.1038/sj.emboj.7601774.
Arif, A., Yao, P., Terenzi, F., Jia, J., Ray, P. S., & Fox, P. L. (2018). The GAIT translational control system. Wiley Interdisciplinary Reviews RNA, 9(2), e1441. https://doi.org/10.1002/wrna.1441.
Ostareck, D. H., Ostareck-Lederer, A., Wilm, M., Thiele, B. J., Mann, M., & Hentze, M. W. (1997). mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89(4), 597–606. https://doi.org/10.1016/s0092-8674(00)80241-x.
Friend, K., Campbell, Z. T., Cooke, A., Kroll-Conner, P., Wickens, M. P., & Kimble, J. (2012). A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nature Structural & Molecular Biology, 19(2), 176–183. https://doi.org/10.1038/nsmb.2214.
Waerner, T., Alacakaptan, M., Tamir, I., Oberauer, R., Gal, A., Brabletz, T., Schreiber, M., Jechlinger, M., & Beug, H. (2006). ILEI: A cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell, 10(3), 227–239. https://doi.org/10.1016/j.ccr.2006.07.020.
Chaudhury, A., Hussey, G. S., Ray, P. S., Jin, G., Fox, P. L., & Howe, P. H. (2010). TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nature Cell Biology, 12(3), 286–293. https://doi.org/10.1038/ncb2029.
Hussey, G. S., Chaudhury, A., Dawson, A. E., Lindner, D. J., Knudsen, C. R., Wilce, M. C., et al. (2011). Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Molecular Cell, 41(4), 419–431. https://doi.org/10.1016/j.molcel.2011.02.003.
Vicario, A., Colliva, A., Ratti, A., Davidovic, L., Baj, G., Gricman, Ł., Colombrita, C., Pallavicini, A., Jones, K. R., Bardoni, B., & Tongiorgi, E. (2015). Dendritic targeting of short and long 3' UTR BDNF mRNA is regulated by BDNF or NT-3 and distinct sets of RNA-binding proteins. Frontiers in Molecular Neuroscience, 8, 62. https://doi.org/10.3389/fnmol.2015.00062.
Melendez, J., Grogg, M., & Zheng, Y. (2011). Signaling role of Cdc42 in regulating mammalian physiology. The Journal of Biological Chemistry, 286(4), 2375–2381. https://doi.org/10.1074/jbc.R110.200329.
Ciolli Mattioli, C., Rom, A., Franke, V., Imami, K., Arrey, G., Terne, M., Woehler, A., Akalin, A., Ulitsky, I., & Chekulaeva, M. (2019). Alternative 3' UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Research, 47(5), 2560–2573. https://doi.org/10.1093/nar/gky1270.
Katz, Z. B., Wells, A. L., Park, H. Y., Wu, B., Shenoy, S. M., & Singer, R. H. (2012). β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes & Development, 26(17), 1885–1890. https://doi.org/10.1101/gad.190413.112.
Rodriguez, A. J., Shenoy, S. M., Singer, R. H., & Condeelis, J. (2006). Visualization of mRNA translation in living cells. The Journal of Cell Biology, 175(1), 67–76. https://doi.org/10.1083/jcb.200512137.
Gutierrez, N., Eromobor, I., Petrie, R. J., Vedula, P., Cruz, L., & Rodriguez, A. J. (2014). The β-actin mRNA zipcode regulates epithelial adherens junction assembly but not maintenance. RNA, 20(5), 689–701. https://doi.org/10.1261/rna.043208.113.
Kislauskis, E. H., Zhu, X., & Singer, R. H. (1994). Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. The Journal of Cell Biology, 127(2), 441–451. https://doi.org/10.1083/jcb.127.2.441.
Hüttelmaier, S., Zenklusen, D., Lederer, M., Dictenberg, J., Lorenz, M., Meng, X., Bassell, G. J., Condeelis, J., & Singer, R. H. (2005). Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature, 438(7067), 512–515. https://doi.org/10.1038/nature04115.
Gu, W., Pan, F., & Singer, R. H. (2009). Blocking beta-catenin binding to the ZBP1 promoter represses ZBP1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. Journal of Cell Science, 122(Pt 11), 1895–1905. https://doi.org/10.1242/jcs.045278.
Gu, W., Katz, Z., Wu, B., Park, H. Y., Li, D., Lin, S., Wells, A. L., & Singer, R. H. (2012). Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. Journal of Cell Science, 125(Pt 1), 81–91. https://doi.org/10.1242/jcs.086132.
Nwokafor, C. U., Sellers, R. S., & Singer, R. H. (2016). IMP1, an mRNA binding protein that reduces the metastatic potential of breast cancer in a mouse model. Oncotarget, 7(45), 72662–72671. https://doi.org/10.18632/oncotarget.12083.
Niedner, A., Edelmann, F. T., & Niessing, D. (2014). Of social molecules: The interactive assembly of ASH1 mRNA-transport complexes in yeast. RNA Biology, 11(8), 998–1009. https://doi.org/10.4161/rna.29946.
Martin, K. C., & Ephrussi, A. (2009). mRNA localization: Gene expression in the spatial dimension. Cell, 136(4), 719–730. https://doi.org/10.1016/j.cell.2009.01.044.
Mili, S., Moissoglu, K., & Macara, I. G. (2008). Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature, 453(7191), 115–119. https://doi.org/10.1038/nature06888.
Wong, H. H., Lin, J. Q., Ströhl, F., Roque, C. G., Cioni, J. M., Cagnetta, R., et al. (2017). RNA docking and local translation regulate site-specific axon remodeling in vivo. Neuron, 95(4), 852–868.e8. https://doi.org/10.1016/j.neuron.2017.07.016.
Preitner, N., Quan, J., Nowakowski, D. W., Hancock, M. L., Shi, J., Tcherkezian, J., Young-Pearse, T. L., & Flanagan, J. G. (2014). APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell, 158(2), 368–382. https://doi.org/10.1016/j.cell.2014.05.042.
Bergsten, S. E., & Gavis, E. R. (1999). Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development, 126(4), 659–669.
Zaessinger, S., Busseau, I., & Simonelig, M. (2006). Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development, 133(22), 4573–4583. https://doi.org/10.1242/dev.02649.
Berkovits, B. D., & Mayr, C. (2015). Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature, 522(7556), 363–367. https://doi.org/10.1038/nature14321.
Jaiswal, S., Jamieson, C. H., Pang, W. W., Park, C. Y., Chao, M. P., Majeti, R., et al. (2009). CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell, 138(2), 271–285. https://doi.org/10.1016/j.cell.2009.05.046.
Ma, W., & Mayr, C. (2018). A membraneless organelle associated with the endoplasmic reticulum enables 3'UTR-mediated protein-protein interactions. Cell, 175(6), 1492–1506.e19. https://doi.org/10.1016/j.cell.2018.10.007.
Grabowski, P. J., Seiler, S. R., & Sharp, P. A. (1985). A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell, 42(1), 345–353. https://doi.org/10.1016/s0092-8674(85)80130-6.
Berget, S. M., Moore, C., & Sharp, P. A. (1977). Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proceedings of the National Academy of Sciences of the United States of America, 74(8), 3171–3175. https://doi.org/10.1073/pnas.74.8.3171.
Chow, L. T., Gelinas, R. E., Broker, T. R., & Roberts, R. J. (1977). An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell, 12(1), 1–8. https://doi.org/10.1016/0092-8674(77)90180-5.
Fica, S. M., & Nagai, K. (2017). Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nature Structural & Molecular Biology, 24(10), 791–799. https://doi.org/10.1038/nsmb.3463.
Climente-González, H., Porta-Pardo, E., Godzik, A., & Eyras, E. (2017). The functional impact of alternative splicing in Cancer. Cell Reports, 20(9), 2215–2226. https://doi.org/10.1016/j.celrep.2017.08.012.
Hoyos, L. E., & Abdel-Wahab, O. (2018). Cancer-specific splicing changes and the potential for splicing-derived neoantigens. Cancer Cell, 34(2), 181–183. https://doi.org/10.1016/j.ccell.2018.07.008.
Anczuków, O., & Krainer, A. R. (2016). Splicing-factor alterations in cancers. RNA, 22(9), 1285–1301. https://doi.org/10.1261/rna.057919.116.
Anczuków, O., Rosenberg, A. Z., Akerman, M., Das, S., Zhan, L., Karni, R., Muthuswamy, S. K., & Krainer, A. R. (2012). The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nature Structural & Molecular Biology, 19(2), 220–228. https://doi.org/10.1038/nsmb.2207.
Karni, R., de Stanchina, E., Lowe, S. W., Sinha, R., Mu, D., & Krainer, A. R. (2007). The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature Structural & Molecular Biology, 14(3), 185–193. https://doi.org/10.1038/nsmb1209.
Ghigna, C., Giordano, S., Shen, H., Benvenuto, F., Castiglioni, F., Comoglio, P. M., Green, M. R., Riva, S., & Biamonti, G. (2005). Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Molecular Cell, 20(6), 881–890. https://doi.org/10.1016/j.molcel.2005.10.026.
Ge, K., DuHadaway, J., Du, W., Herlyn, M., Rodeck, U., & Prendergast, G. C. (1999). Mechanism for elimination of a tumor suppressor: Aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proceedings of the National Academy of Sciences of the United States of America, 96(17), 9689–9694. https://doi.org/10.1073/pnas.96.17.9689.
Scheper, G. C., Parra, J. L., Wilson, M., Van Kollenburg, B., Vertegaal, A. C., Han, Z. G., et al. (2003). The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Molecular and Cellular Biology, 23(16), 5692–5705. https://doi.org/10.1128/mcb.23.16.5692-5705.2003.
Chen, L., Luo, C., Shen, L., Liu, Y., Wang, Q., Zhang, C., Guo, R., Zhang, Y., Xie, Z., Wei, N., Wu, W., Han, J., & Feng, Y. (2017). SRSF1 prevents DNA damage and promotes tumorigenesis through regulation of DBF4B pre-mRNA splicing. Cell Reports, 21(12), 3406–3413. https://doi.org/10.1016/j.celrep.2017.11.091.
Dvinge, H., Kim, E., Abdel-Wahab, O., & Bradley, R. K. (2016). RNA splicing factors as oncoproteins and tumour suppressors. Nature Reviews. Cancer, 16(7), 413–430. https://doi.org/10.1038/nrc.2016.51.
Urbanski, L. M., Leclair, N., & Anczuków, O. (2018). Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdisciplinary Reviews RNA, 9(4), e1476. https://doi.org/10.1002/wrna.1476.
Obeng, E. A., Stewart, C., & Abdel-Wahab, O. (2019). Altered RNA processing in cancer pathogenesis and therapy. Cancer Discovery, 9(11), 1493–1510. https://doi.org/10.1158/2159-8290.CD-19-0399.
Dutertre, M., Chakrama, F. Z., Combe, E., Desmet, F. O., Mortada, H., Polay Espinoza, M., Gratadou, L., & Auboeuf, D. (2014). A recently evolved class of alternative 3′-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nature Communications, 5, 3395. https://doi.org/10.1038/ncomms4395.
Vorlová, S., Rocco, G., Lefave, C. V., Jodelka, F. M., Hess, K., Hastings, M. L., et al. (2011). Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Molecular Cell, 43(6), 927–939. https://doi.org/10.1016/j.molcel.2011.08.009.
Lee, S. H., Singh, I., Tisdale, S., Abdel-Wahab, O., Leslie, C. S., & Mayr, C. (2018). Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature, 561(7721), 127–131. https://doi.org/10.1038/s41586-018-0465-8.
Graber, J. H., Nazeer, F. I., Yeh, P. C., Kuehner, J. N., Borikar, S., Hoskinson, D., & Moore, C. L. (2013). DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Research, 23(10), 1690–1703. https://doi.org/10.1101/gr.144964.112.
Davidson, L., Muniz, L., & West, S. (2014). 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes & Development, 28(4), 342–356. https://doi.org/10.1101/gad.231274.113.
Dubbury, S. J., Boutz, P. L., & Sharp, P. A. (2018). CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature, 564(7734), 141–145. https://doi.org/10.1038/s41586-018-0758-y.
Quereda, V., Bayle, S., Vena, F., Frydman, S. M., Monastyrskyi, A., Roush, W. R., et al. (2019). Therapeutic targeting of CDK12/CDK13 in triple-negative breast cancer. Cancer Cell, 36(5), 545–558.e7. https://doi.org/10.1016/j.ccell.2019.09.004.
Tian, B., Hu, J., Zhang, H., & Lutz, C. S. (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Research, 33(1), 201–212. https://doi.org/10.1093/nar/gki158.
Venters, C. C., Oh, J. M., Di, C., So, B. R., & Dreyfuss, G. (2019). U1 snRNP telescripting: Suppression of premature transcription termination in introns as a new layer of gene regulation. Cold Spring Harbor Perspectives in Biology, 11(2). https://doi.org/10.1101/cshperspect.a032235.
Tian, B., & Manley, J. L. (2017). Alternative polyadenylation of mRNA precursors. Nature Reviews. Molecular Cell Biology, 18(1), 18–30. https://doi.org/10.1038/nrm.2016.116.
Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A., & Burge, C. B. (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science, 320(5883), 1643–1647. https://doi.org/10.1126/science.1155390.
Mayr, C., & Bartel, D. P. (2009). Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell, 138(4), 673–684. https://doi.org/10.1016/j.cell.2009.06.016.
Rosenwald, A., Wright, G., Wiestner, A., Chan, W. C., Connors, J. M., Campo, E., Gascoyne, R. D., Grogan, T. M., Muller-Hermelink, H. K., Smeland, E. B., Chiorazzi, M., Giltnane, J. M., Hurt, E. M., Zhao, H., Averett, L., Henrickson, S., Yang, L., Powell, J., Wilson, W. H., Jaffe, E. S., Simon, R., Klausner, R. D., Montserrat, E., Bosch, F., Greiner, T. C., Weisenburger, D. D., Sanger, W. G., Dave, B. J., Lynch, J. C., Vose, J., Armitage, J. O., Fisher, R. I., Miller, T. P., LeBlanc, M., Ott, G., Kvaloy, S., Holte, H., Delabie, J., & Staudt, L. M. (2003). The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell, 3(2), 185–197.
Xia, Z., Donehower, L. A., Cooper, T. A., Neilson, J. R., Wheeler, D. A., Wagner, E. J., & Li, W. (2014). Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3'-UTR landscape across seven tumour types. Nature Communications, 5, 5274. https://doi.org/10.1038/ncomms6274.
Begik, O., Oyken, M., Cinkilli Alican, T., Can, T., & Erson-Bensan, A. E. (2017). Alternative polyadenylation patterns for novel gene discovery and classification in Cancer. Neoplasia, 19(7), 574–582. https://doi.org/10.1016/j.neo.2017.04.008.
Akman, B. H., Can, T., & Erson-Bensan, A. E. (2012). Estrogen-induced upregulation and 3'-UTR shortening of CDC6. Nucleic Acids Research, 40(21), 10679–10688. https://doi.org/10.1093/nar/gks855.
Akman, H. B., Oyken, M., Tuncer, T., Can, T., & Erson-Bensan, A. E. (2015). 3 ' UTR shortening and EGF signaling: Implications for breast cancer. Human Molecular Genetics, 24(24), 6910–6920. https://doi.org/10.1093/hmg/ddv391.
Miles, W. O., Lembo, A., Volorio, A., Brachtel, E., Tian, B., Sgroi, D., Provero, P., & Dyson, N. (2016). Alternative polyadenylation in triple-negative breast tumors allows NRAS and c-JUN to bypass PUMILIO posttranscriptional regulation. Cancer Research, 76(24), 7231–7241. https://doi.org/10.1158/0008-5472.CAN-16-0844.
Chen, M., Lyu, G., Han, M., Nie, H., Shen, T., Chen, W., Niu, Y., Song, Y., Li, X., Li, H., Chen, X., Wang, Z., Xia, Z., Li, W., Tian, X. L., Ding, C., Gu, J., Zheng, Y., Liu, X., Hu, J., Wei, G., Tao, W., & Ni, T. (2018). 3' UTR lengthening as a novel mechanism in regulating cellular senescence. Genome Research, 28, 285–294. https://doi.org/10.1101/gr.224451.117.
Jia, Q., Nie, H., Yu, P., Xie, B., Wang, C., Yang, F., et al. (2019). HNRNPA1-mediated 3' UTR length changes of HN1 contributes to cancer- and senescence-associated phenotypes. Aging (Albany NY), 11(13), 4407–4437. https://doi.org/10.18632/aging.102060.
Matallanas, D., Birtwistle, M., Romano, D., Zebisch, A., Rauch, J., von Kriegsheim, A., & Kolch, W. (2011). Raf family kinases: Old dogs have learned new tricks. Genes & Cancer, 2(3), 232–260. https://doi.org/10.1177/1947601911407323.
Cantwell-Dorris, E. R., O'Leary, J. J., & Sheils, O. M. (2011). BRAFV600E: Implications for carcinogenesis and molecular therapy. Molecular Cancer Therapeutics, 10(3), 385–394. https://doi.org/10.1158/1535-7163.MCT-10-0799.
Kim, A., & Cohen, M. S. (2016). The discovery of vemurafenib for the treatment of BRAF-mutated metastatic melanoma. Expert Opinion on Drug Discovery, 11(9), 907–916. https://doi.org/10.1080/17460441.2016.1201057.
Marranci, A., Jiang, Z., Vitiello, M., Guzzolino, E., Comelli, L., Sarti, S., Lubrano, S., Franchin, C., Echevarría-Vargas, I., Tuccoli, A., Mercatanti, A., Evangelista, M., Sportoletti, P., Cozza, G., Luzi, E., Capobianco, E., Villanueva, J., Arrigoni, G., Signore, G., Rocchiccioli, S., Pitto, L., Tsinoremas, N., & Poliseno, L. (2017). The landscape of BRAF transcript and protein variants in human cancer. Molecular Cancer, 16(1), 85. https://doi.org/10.1186/s12943-017-0645-4.
Marranci, A., Tuccoli, A., Vitiello, M., Mercoledi, E., Sarti, S., Lubrano, S., Evangelista, M., Fogli, A., Valdes, C., Russo, F., Monte, M. D., Caligo, M. A., Pellegrini, M., Capobianco, E., Tsinoremas, N., & Poliseno, L. (2015). Identification of BRAF 3'UTR isoforms in melanoma. The Journal of Investigative Dermatology, 135(6), 1694–1697. https://doi.org/10.1038/jid.2015.47.
Pardoll, D. M. (2012). The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews. Cancer, 12(4), 252–264. https://doi.org/10.1038/nrc3239.
Shi, Y. (2018). Regulatory mechanisms of PD-L1 expression in cancer cells. Cancer Immunology, Immunotherapy, 67(10), 1481–1489. https://doi.org/10.1007/s00262-018-2226-9.
Kataoka, K., Shiraishi, Y., Takeda, Y., Sakata, S., Matsumoto, M., Nagano, S., Maeda, T., Nagata, Y., Kitanaka, A., Mizuno, S., Tanaka, H., Chiba, K., Ito, S., Watatani, Y., Kakiuchi, N., Suzuki, H., Yoshizato, T., Yoshida, K., Sanada, M., Itonaga, H., Imaizumi, Y., Totoki, Y., Munakata, W., Nakamura, H., Hama, N., Shide, K., Kubuki, Y., Hidaka, T., Kameda, T., Masuda, K., Minato, N., Kashiwase, K., Izutsu, K., Takaori-Kondo, A., Miyazaki, Y., Takahashi, S., Shibata, T., Kawamoto, H., Akatsuka, Y., Shimoda, K., Takeuchi, K., Seya, T., Miyano, S., & Ogawa, S. (2016). Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature, 534(7607), 402–406. https://doi.org/10.1038/nature18294.
Hassounah, N. B., Malladi, V. S., Huang, Y., Freeman, S. S., Beauchamp, E. M., Koyama, S., Souders, N., Martin, S., Dranoff, G., Wong, K. K., Pedamallu, C. S., Hammerman, P. S., & Akbay, E. A. (2019). Identification and characterization of an alternative cancer-derived PD-L1 splice variant. Cancer Immunology, Immunotherapy, 68(3), 407–420. https://doi.org/10.1007/s00262-018-2284-z.
Mahoney, K. M., Shukla, S. A., Patsoukis, N., Chaudhri, A., Browne, E. P., Arazi, A., Eisenhaure, T. M., Pendergraft III, W. F., Hua, P., Pham, H. C., Bu, X., Zhu, B., Hacohen, N., Fritsch, E. F., Boussiotis, V. A., Wu, C. J., & Freeman, G. J. (2019). A secreted PD-L1 splice variant that covalently dimerizes and mediates immunosuppression. Cancer Immunology, Immunotherapy, 68(3), 421–432. https://doi.org/10.1007/s00262-018-2282-1.
Audrito, V., Serra, S., Stingi, A., Orso, F., Gaudino, F., Bologna, C., et al. (2017). PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget, 8(9), 15894–15911. https://doi.org/10.18632/oncotarget.15213.
Xie, W. B., Liang, L. H., Wu, K. G., Wang, L. X., He, X., Song, C., Wang, Y. Q., & Li, Y. H. (2018). MiR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cellular Physiology and Biochemistry, 46(2), 654–663. https://doi.org/10.1159/000488634.
Puente, X. S., Beà, S., Valdés-Mas, R., Villamor, N., Gutiérrez-Abril, J., Martín-Subero, J. I., Munar, M., Rubio-Pérez, C., Jares, P., Aymerich, M., Baumann, T., Beekman, R., Belver, L., Carrio, A., Castellano, G., Clot, G., Colado, E., Colomer, D., Costa, D., Delgado, J., Enjuanes, A., Estivill, X., Ferrando, A. A., Gelpí, J. L., González, B., González, S., González, M., Gut, M., Hernández-Rivas, J. M., López-Guerra, M., Martín-García, D., Navarro, A., Nicolás, P., Orozco, M., Payer, Á. R., Pinyol, M., Pisano, D. G., Puente, D. A., Queirós, A. C., Quesada, V., Romeo-Casabona, C. M., Royo, C., Royo, R., Rozman, M., Russiñol, N., Salaverría, I., Stamatopoulos, K., Stunnenberg, H. G., Tamborero, D., Terol, M. J., Valencia, A., López-Bigas, N., Torrents, D., Gut, I., López-Guillermo, A., López-Otín, C., & Campo, E. (2015). Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature, 526(7574), 519–524. https://doi.org/10.1038/nature14666.
Wang, W., Sun, J., Li, F., Li, R., Gu, Y., Liu, C., Yang, P., Zhu, M., Chen, L., Tian, W., Zhou, H., Mao, Y., Zhang, L., Jiang, J., Wu, C., Hua, D., Chen, W., Lu, B., Ju, J., & Zhang, X. (2012). A frequent somatic mutation in CD274 3'-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Human Mutation, 33(3), 480–484. https://doi.org/10.1002/humu.22014.
Desrosiers, R., Friderici, K., & Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America, 71(10), 3971–3975. https://doi.org/10.1073/pnas.71.10.3971.
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., Cesarkas, K., Jacob-Hirsch, J., Amariglio, N., Kupiec, M., Sorek, R., & Rechavi, G. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 485(7397), 201–206. https://doi.org/10.1038/nature11112.
Bushkin, G. G., Pincus, D., Morgan, J. T., Richardson, K., Lewis, C., Chan, S. H., Bartel, D. P., & Fink, G. R. (2019). m6A modification of a 3' UTR site reduces RME1 mRNA levels to promote meiosis. Nature Communications, 10(1), 3414. https://doi.org/10.1038/s41467-019-11232-7.
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., He, X., & Semenza, G. L. (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proceedings of the National Academy of Sciences of the United States of America, 113(14), E2047–E2056. https://doi.org/10.1073/pnas.1602883113.
Erson-Bensan, A. E., & Can, T. (2016). Alternative Polyadenylation: Another foe in cancer. Molecular Cancer Research. https://doi.org/10.1158/1541-7786.MCR-15-0489.
Gruber, A. J., & Zavolan, M. (2019). Alternative cleavage and polyadenylation in health and disease. Nature Reviews. Genetics, 20(10), 599–614. https://doi.org/10.1038/s41576-019-0145-z.
Shulman, E. D., & Elkon, R. (2019). Cell-type-specific analysis of alternative polyadenylation using single-cell transcriptomics data. Nucleic Acids Research, 47(19), 10027–10039. https://doi.org/10.1093/nar/gkz781.
Steber, H. S., Gallante, C., O'Brien, S., Chiu, P. L., & Mangone, M. (2019). The C. elegans 3' UTRome v2 resource for studying mRNA cleavage and polyadenylation, 3'-UTR biology, and miRNA targeting. Genome Research, 29(12), 2104–2116. https://doi.org/10.1101/gr.254839.119.
Acknowledgments
The author sincerely apologizes to colleagues whose important work could not be cited in this review article because of space limitations, and thanks the laboratory members for critical reading of the manuscript. Funding for our laboratory’s work on 3′UTRs was from TUBITAK and METU.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Erson-Bensan, A.E. RNA-biology ruling cancer progression? Focus on 3′UTRs and splicing. Cancer Metastasis Rev 39, 887–901 (2020). https://doi.org/10.1007/s10555-020-09884-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-020-09884-9