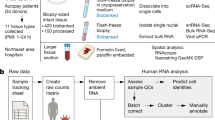
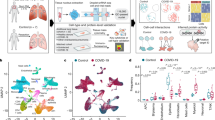
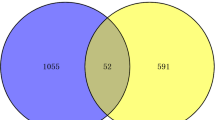
Abstract
Severe COVID-19 is a systemic disorder involving excessive inflammatory response, metabolic dysfunction, multi-organ damage, and several clinical features. Here, we performed a transcriptome meta-analysis investigating genes and molecular mechanisms related to COVID-19 severity and outcomes. First, transcriptomic data of cellular models of SARS-CoV-2 infection were compiled to understand the first response to the infection. Then, transcriptomic data from lung autopsies of patients deceased due to COVID-19 were compiled to analyze altered genes of damaged lung tissue. These analyses were followed by functional enrichment analyses and gene–phenotype association. A biological network was constructed using the disturbed genes in the lung autopsy meta-analysis. Central genes were defined considering closeness and betweenness centrality degrees. A sub-network phenotype–gene interaction analysis was performed. The meta-analysis of cellular models found genes mainly associated with cytokine signaling and other pathogen response pathways. The meta-analysis of lung autopsy tissue found genes associated with coagulopathy, lung fibrosis, multi-organ damage, and long COVID-19. Only genes DNAH9 and FAM216B were found perturbed in both meta-analyses. BLNK, FABP4, GRIA1, ATF3, TREM2, TPPP, TPPP3, FOS, ALB, JUNB, LMNA, ADRB2, PPARG, TNNC1, and EGR1 were identified as central elements among perturbed genes in lung autopsy and were found associated with several clinical features of severe COVID-19. Central elements were suggested as interesting targets to investigate the relation with features of COVID-19 severity, such as coagulopathy, lung fibrosis, and organ damage.







Similar content being viewed by others
References
Ackermann M, Verleden SE, Kuehnel M et al (2020) Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID-19. N Engl J Med 383:120–128. https://doi.org/10.1056/NEJMOA2015432
Adivitiya KMS, Chakraborty S et al (2021) Mucociliary respiratory epithelium integrity in molecular defense and susceptibility to pulmonary viral infections. Biology (Basel) 10:95. https://doi.org/10.3390/biology10020095
Afgan E, Baker D, Batut B et al (2018) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. https://doi.org/10.1093/NAR/GKY379
Aguiar D, Lobrinus JA, Schibler M et al (2020) Inside the lungs of COVID-19 disease. Int J Legal Med 134:1271–1274. https://doi.org/10.1007/S00414-020-02318-9
Alimadadi A, Munroe PB, Joe B, Cheng X (2020) Meta-analysis of dilated cardiomyopathy using cardiac RNA-Seq transcriptomic datasets. Genes (Basel) 11(1):60. https://doi.org/10.3390/genes11010060
Alsamman AM, Zayed H (2020) The transcriptomic profiling of SARS-CoV-2 compared to SARS, MERS, EBOV, and H1N1. PLoS ONE 15:e0243270. https://doi.org/10.1371/journal.pone.0243270
Andrews S (2023) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 1 June 2023.
Asakura H, Ogawa H (2021) COVID-19-associated coagulopathy and disseminated intravascular coagulation. Int J Hematol 113:45–57. https://doi.org/10.1007/s12185-020-03029-y
Asano T, Boisson B, Onodi F et al (2021) X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci Immunol 6(62):eabl4348. https://doi.org/10.1126/sciimmunol.abl4348
Bortolotti D, Gentili V, Rizzo S et al (2021) TLR3 and TLR7 RNA sensor activation during SARS-CoV-2 infection. Microorganisms 9(9):1820. https://doi.org/10.3390/MICROORGANISMS9091820
Budhraja A, Basu A, Gheware A et al (2022) Molecular signature of postmortem lung tissue from COVID-19 patients suggests distinct trajectories driving mortality. Dis Model Mech 15(5):dmm049572. https://doi.org/10.1242/dmm.049572
Celik E, Nelles C, Kottlors J et al (2022) Quantitative determination of pulmonary emphysema in follow-up LD-CTs of patients with COVID-19 infection. PLoS ONE 17:e0263261. https://doi.org/10.1371/journal.pone.0263261
Cheng Y, Luo R, Wang K et al (2020) Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int 97:829–838. https://doi.org/10.1016/j.kint.2020.03.005
Chinetti G, Fruchart J-C, Staels B (2000) Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm Res 49:497–505. https://doi.org/10.1007/s000110050622
De Vriese AS, Sethi S, Van Praet J et al (2015) Kidney disease caused by dysregulation of the complement alternative pathway: an etiologic approach. J Am Soc Nephrol 26:2917–2929. https://doi.org/10.1681/ASN.2015020184
Desai N, Neyaz A, Szabolcs A et al (2020) Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun 11:6319. https://doi.org/10.1038/s41467-020-20139-7
Diamond MS, Kanneganti TD (2022) Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol 23:165–176. https://doi.org/10.1038/S41590-021-01091-0
Fabbri L, Moss S, Khan FA et al (2022) Parenchymal lung abnormalities following hospitalisation for COVID-19 and viral pneumonitis: a systematic review and meta-analysis. Thorax 78(2):191–201. https://doi.org/10.1136/thoraxjnl-2021-218275
Fassad MR, Shoemark A, Legendre M et al (2018) Mutations in outer dynein arm heavy chain DNAH9 cause motile cilia defects and situs inversus. Am J Hum Genet 103:984–994. https://doi.org/10.1016/j.ajhg.2018.10.016
Fearn A (2015) Complement activation in progressive renal disease. World J Nephrol 4:31. https://doi.org/10.5527/wjn.v4.i1.31
Fliegauf M, Olbrich H, Horvath J et al (2005) Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med 171:1343–1349. https://doi.org/10.1164/rccm.200411-1583OC
Guo C, Li B, Ma H et al (2020) (2020) Single-cell analysis of two severe COVID-19 patients reveals a monocyte-associated and tocilizumab-responding cytokine storm. Nat Commun 111(11):1–11. https://doi.org/10.1038/s41467-020-17834-w
Han Y, Yang L, Duan X et al (2020) Identification of candidate COVID-19 therapeutics using hPSC-derived lung organoids. bioRxiv (Preprint). https://doi.org/10.1101/2020.05.05.079095
Han X, Fan Y, Alwalid O et al (2021a) Six-month follow-up chest CT findings after severe COVID-19 pneumonia. Radiology 299:E177–E186. https://doi.org/10.1148/radiol.2021203153
Han Y, Duan X, Yang L et al (2021b) Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589:270–275. https://doi.org/10.1038/S41586-020-2901-9
Harb H, Benamar M, Lai PS et al (2021) Notch4 signaling limits regulatory T-cell-mediated tissue repair and promotes severe lung inflammation in viral infections. Immunity 54:1186-1199.e7. https://doi.org/10.1016/j.immuni.2021.04.002
Hartenian E, Nandakumar D, Lari A et al (2020) The molecular virology of coronaviruses. J Biol Chem 295:12910–12934. https://doi.org/10.1074/JBC.REV120.013930
Huang WJ, Tang XX (2021) Virus infection induced pulmonary fibrosis. J Transl Med 19:496. https://doi.org/10.1186/s12967-021-03159-9
Huang C, Wang Y, Li X et al (2020a) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. https://doi.org/10.1016/S0140-6736(20)30183-5
Huang J, Hume AJ, Abo KM et al (2020b) SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell 27:962-973.e7. https://doi.org/10.1016/J.STEM.2020.09.013
Huang J, Wang Y, Zha Y et al (2022) Transcriptome analysis reveals hub genes regulating autophagy in patients with severe COVID-19. Front Genet 13:908826. https://doi.org/10.3389/fgene.2022.908826
Iverson E, Kaler L, Agostino EL et al (2020) Leveraging 3D model systems to understand viral interactions with the respiratory mucosa. Viruses 12:1425. https://doi.org/10.3390/v12121425
Jaffe AE, Hyde T, Kleinman J et al (2015) Practical impacts of genomic data “cleaning” on biological discovery using surrogate variable analysis. BMC Bioinform 16:372. https://doi.org/10.1186/S12859-015-0808-5
Kowalski TW, Lord VO, Sgarioni E et al (2022) Transcriptome meta-analysis of valproic acid exposure in human embryonic stem cells. Eur Neuropsychopharmacol 60:76–88. https://doi.org/10.1016/j.euroneuro.2022.04.008
Lamers MM, van der Vaart J, Knoops K et al (2021) An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J 40:e105912. https://doi.org/10.15252/embj.2020105912
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25. https://doi.org/10.1186/GB-2009-10-3-R25
Leek JT, Johnson WE, Parker HS et al (2022) sva: surrogate variable analysis
Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration (2011) The sequence read archive. Nucleic Acids Res 39:D19–D21. https://doi.org/10.1093/NAR/GKQ1019
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/BIOINFORMATICS/BTT656
Lodigiani C, Iapichino G, Carenzo L et al (2020) Venous and arterial thromboembolic complications in COVID-19 patients admitted to an academic hospital in Milan, Italy. Thromb Res 191:9–14. https://doi.org/10.1016/j.thromres.2020.04.024
Luo W, Brouwer C (2013) Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29:1830–1831. https://doi.org/10.1093/bioinformatics/btt285
Luo W, Friedman MS, Shedden K et al (2009) GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinform 10:161. https://doi.org/10.1186/1471-2105-10-161
Maleknia S, Tavassolifar MJ, Mottaghitalab F et al (2022) Identifying novel host-based diagnostic biomarker panels for COVID-19: a whole-blood/nasopharyngeal transcriptome meta-analysis. Mol Med 28:86. https://doi.org/10.1186/s10020-022-00513-5
Mathew D, Giles JR, Baxter AE et al (2020) Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369(6508):eabc8511. https://doi.org/10.1126/SCIENCE.ABC8511
Maxwell AJ, Ding J, You Y et al (2021) Identification of key signaling pathways induced by SARS-CoV2 that underlie thrombosis and vascular injury in COVID-19 patients. J Leukoc Biol 109:35–47. https://doi.org/10.1002/JLB.4COVR0920-552RR
Mulay A, Konda B, Garcia G et al (2021) SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep 35:109055. https://doi.org/10.1016/j.celrep.2021.109055
Mulchandani R, Lyngdoh T, Kakkar AK (2021) Deciphering the COVID-19 cytokine storm: systematic review and meta-analysis. Eur J Clin Investig 51(1):e13429. https://doi.org/10.1111/ECI.13429
Østergaard L (2021) SARS CoV-2 related microvascular damage and symptoms during and after COVID-19: consequences of capillary transit-time changes, tissue hypoxia and inflammation. Physiol Rep 9(3):e14726. https://doi.org/10.14814/phy2.14726
Pairo-Castineira E, Clohisey S, Klaric L et al (2021) Genetic mechanisms of critical illness in COVID-19. Nature 591:92–98. https://doi.org/10.1038/s41586-020-03065-y
Perico L, Benigni A, Casiraghi F et al (2021) Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol 17:46–64. https://doi.org/10.1038/S41581-020-00357-4
Pernazza A, Mancini M, Rullo E et al (2020) Early histologic findings of pulmonary SARS-CoV-2 infection detected in a surgical specimen. Virchows Arch 477:743–748. https://doi.org/10.1007/S00428-020-02829-1
Prada C, Lima D, Nakaya H (2022) MetaVolcanoR: gene expression meta-analysis visualization tool
Qi F, Qian S, Zhang S, Zhang Z (2020) Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun 526:135–140. https://doi.org/10.1016/j.bbrc.2020.03.044
Qin C, Zhou L, Hu Z et al (2020) Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis 71:762–768. https://doi.org/10.1093/cid/ciaa248
Qu J, Zhu H-H, Huang X-J et al (2021) Abnormal indexes of liver and kidney injury markers predict severity in COVID-19 patients. Infect Drug Resist 14:3029–3040. https://doi.org/10.2147/IDR.S321915
Rau A, Marot G, Jaffrézic F (2014) Differential meta-analysis of RNA-Seq data from multiple studies. BMC Bioinform 15:91. https://doi.org/10.1186/1471-2105-15-91
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/BIOINFORMATICS/BTP616
Rodrigues TS, de Sá KSG, Ishimoto AY et al (2021) Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med 218(3):e20201707. https://doi.org/10.1084/JEM.20201707
Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E et al (2020) Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis 34:101623. https://doi.org/10.1016/J.TMAID.2020.101623
Rommasi F, Nasiri MJ, Mirsaeidi M (2022) Immunomodulatory agents for COVID-19 treatment: possible mechanism of action and immunopathology features. Mol Cell Biochem 477:711–726. https://doi.org/10.1007/s11010-021-04325-9
Salem ML, Eltoukhy MM, Shalaby RE et al (2023) COVID-19 severity shifts the cytokine milieu toward a proinflammatory state in Egyptian patients: a cross-sectional study. J Interferon Cytokine Res. https://doi.org/10.1089/jir.2023.0029
Sano E, Suzuki T, Hashimoto R et al (2022) Cell response analysis in SARS-CoV-2 infected bronchial organoids. Commun Biol 5:516. https://doi.org/10.1038/s42003-022-03499-2
Scardoni G, Petterlini M, Laudanna C (2009) Analyzing biological network parameters with CentiScaPe. Bioinformatics 25:2857–2859. https://doi.org/10.1093/bioinformatics/btp517
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Silvin A, Chapuis N, Dunsmore G et al (2020) Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 182:1401-1418.e18. https://doi.org/10.1016/J.CELL.2020.08.002
Soul J, Dunn SL, Hardingham TE et al (2016) PhenomeScape: a cytoscape app to identify differentially regulated sub-networks using known disease associations. Bioinformatics 32:3847–3849. https://doi.org/10.1093/bioinformatics/btw545
Stokes EK, Zambrano LD, Anderson KN et al (2020) Coronavirus disease 2019 case surveillance—United States, January 22–May 30, 2020. Morb Mortal Wkly Rep 69:759–765. https://doi.org/10.15585/MMWR.MM6924E2
Szklarczyk D, Gable AL, Nastou KC et al (2021) The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612. https://doi.org/10.1093/nar/gkaa1074
Sweeney TE, Haynes WA, Vallania F et al (2017) Methods to increase reproducibility in differential gene expression via meta-analysis. Nucleic Acids Res 45:e1. https://doi.org/10.1093/nar/gkw797
Tang N, Li D, Wang X, Sun Z (2020) Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 18:844–847. https://doi.org/10.1111/jth.14768
Tang D, Sha Y, Gao Y et al (2021) Novel variants in DNAH9 lead to nonsyndromic severe asthenozoospermia. Reprod Biol Endocrinol 19:27. https://doi.org/10.1186/s12958-021-00709-0
Teuwen LA, Geldhof V, Pasut A, Carmeliet P (2020) COVID-19: the vasculature unleashed. Nat Rev Immunol 207(20):389–391. https://doi.org/10.1038/s41577-020-0343-0
Thorne LG, Reuschl A, Zuliani-Alvarez L et al (2021) SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J 40(15):e107826. https://doi.org/10.15252/EMBJ.2021107826
Tindle C, Fuller M, Fonseca A et al (2021) Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. eLife 10:e66417. https://doi.org/10.7554/eLife.66417
Toro-Domínguez D, Villatoro-Garciá JA, Martorell-Marugán J et al (2021) A survey of gene expression meta-analysis: methods and applications. Brief Bioinform 22:1694–1705. https://doi.org/10.1093/BIB/BBAA019
Torres-Castro R, Vasconcello-Castillo L, Alsina-Restoy X et al (2021) Respiratory function in patients post-infection by COVID-19: a systematic review and meta-analysis. Pulmonology 27:328–337. https://doi.org/10.1016/j.pulmoe.2020.10.013
Tudoran C, Tudoran M, Lazureanu VE et al (2021) Evidence of pulmonary hypertension after SARS-CoV-2 infection in subjects without previous significant cardiovascular pathology. J Clin Med 10:199. https://doi.org/10.3390/jcm10020199
Wan Y-W, Al-Ouran R, Mangleburg CG et al (2020) Meta-analysis of the Alzheimer’s disease human brain transcriptome and functional dissection in mouse models. Cell Rep 32:107908. https://doi.org/10.1016/j.celrep.2020.107908
Winter C, Camarão AAR, Steffen I, Jung K (2022) Network meta-analysis of transcriptome expression changes in different manifestations of dengue virus infection. BMC Genomics 23:165. https://doi.org/10.1186/s12864-022-08390-2
Wu Z, McGoogan JM (2020) Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 323:1239–1242. https://doi.org/10.1001/JAMA.2020.2648
Xiong Y, Liu Y, Cao L et al (2020) Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 9:761–770. https://doi.org/10.1080/22221751.2020.1747363
Ye M, Wysocki J, William J et al (2006) Glomerular localization and expression of angiotensin-converting enzyme 2 and angiotensin-converting enzyme: implications for albuminuria in diabetes. J Am Soc Nephrol 17:3067–3075. https://doi.org/10.1681/ASN.2006050423
Yongzhi X (2021) COVID-19-associated cytokine storm syndrome and diagnostic principles: an old and new Issue. Emerg Microbes Infect 10:266–276. https://doi.org/10.1080/22221751.2021.1884503
Youk J, Kim T, Evans KV et al (2020) Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell 27:905-919.e10. https://doi.org/10.1016/J.STEM.2020.10.004
Yu G, He QY (2016) ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 12:477–479. https://doi.org/10.1039/C5MB00663E
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol 16:284–287. https://doi.org/10.1089/omi.2011.0118
Yu G, Wang LG, Yan GR, He QY (2015) DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31:608–609. https://doi.org/10.1093/BIOINFORMATICS/BTU684
Zhang Q, Bastard P, Liu Z et al (2020) Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370(6515):eabd4570. https://doi.org/10.1126/science.abd4570
Zhou G, Stevenson MM, Geary TG, Xia J (2016) Comprehensive transcriptome meta-analysis to characterize host immune responses in helminth infections. PLoS Negl Trop Dis 10:e0004624. https://doi.org/10.1371/journal.pntd.0004624
Funding
N.A.C. was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Grant 88887.518451/2020-00. T.W.K. was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Grant 150181/2023-0. F.S.L.V. is recipient of a CNPq scholarship, Grant 312960/2021-2.
Author information
Authors and Affiliations
Contributions
Conceptualization: NAC, TWK, MRM, and FSLV; Methodology: NAC, TWK, and MRM; Formal analysis and investigation: NAC, VOL, and TWK; Writing—original draft preparation: NAC and TWK; Writing—review and editing: NAC, VOL, TWK, MRM, and FSLV; Funding acquisition: FSLV; Resources: TWK, MRM, and FSLV; Supervision: TWK, MRM, and FSLV.
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that there is no conflict of interest related to the manuscript. The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2023_10453_MOESM1_ESM.xlsx
Supplementary file1 (XLSX 2 KB) Meta-analysis results of cellular models data. Meta-analysis results of transcriptome data from cellular models of SARS-CoV-2 infections
10528_2023_10453_MOESM2_ESM.xlsx
Supplementary file2 (XLSX 2 KB) DisGeNet enrichment analysis of lung autopsy data. Gene-disease association of perturbed genes from lung autopsy meta-analysis
10528_2023_10453_MOESM3_ESM.xlsx
Supplementary file3 (PDF 2 KB) Gene ontology enrichment analysis of cellular models. Biological processes identified through gene ontology analysis using perturbed genes from cellular models meta-analysis
10528_2023_10453_MOESM4_ESM.xlsx
Supplementary file4 (XLSX 2 KB) Reactome enrichment analysis of cellular models. Reactome pathways identified using perturbed genes from cellular models meta-analysis
10528_2023_10453_MOESM5_ESM.xlsx
Supplementary file5 (XLSX 2 KB) KEGG pathways enrichment analysis of infected cellular models. KEGG enrichment analysis using metaFC values from the meta-analysis of cellular models of infection
10528_2023_10453_MOESM6_ESM.xlsx
Supplementary file6 (XLSX 2 KB) Meta-analysis results of lung autopsy data. Meta-analysis results of transcriptome data from lung autopsy of COVID-19 patients
10528_2023_10453_MOESM7_ESM.xlsx
Supplementary file7 (XLSX 2 KB) Gene ontology enrichment analysis of lung autopsy. Biological processes enriched in gene ontology analysis using perturbed genes from lung autopsy meta-analysis
10528_2023_10453_MOESM9_ESM.xlsx
Supplementary file9 (XLSX 2 KB) Collected datasets from NCBI. Information of selected datasets and samples included in the meta-analysis
10528_2023_10453_MOESM10_ESM.xlsx
Supplementary file10 (XLSX 2 KB) Reactome enrichment analysis of lung autopsy. Reactome pathways identified using perturbed genes from lung autopsy meta-analysis
10528_2023_10453_MOESM11_ESM.xlsx
Supplementary file11 (XLSX 2 KB) KEGG pathways enrichment analysis of COVID-19 lung autopsy. KEGG enrichment analysis using metaFC values from the meta-analysis of lung autopsy
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cadore, N.A., Lord, V.O., Recamonde-Mendoza, M. et al. Meta-analysis of Transcriptomic Data from Lung Autopsy and Cellular Models of SARS-CoV-2 Infection. Biochem Genet 62, 892–914 (2024). https://doi.org/10.1007/s10528-023-10453-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10453-2