Abstract
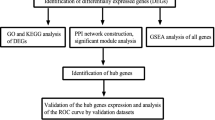
Tubulointerstitial fibrosis is an important pathological feature of diabetic nephropathy that is associated with impaired renal function. However, the mechanism by which fibrosis occurs in diabetic nephropathy is unclear. Differentially expressed genes were identified from transcriptome profiles of renal tissue from diabetic patients and unilateral ureteral obstruction mice and intersected to obtain genes that may be involved in diabetic fibrosis. Biological function analysis and protein–protein interaction network analysis were performed. ROC curve and Pearson correlation analysis between hub genes were performed and glomerular filtration rate estimated. Finally, the RNA levels of hub genes were measured using real-time PCR. A total of 283 genes were identified as potentially involved in diabetic nephropathy fibrosis. TYROBP, CTSS, LCP2, LUM and TLR7 were identified as aberrantly expressed hub genes. Immune cell infiltration analysis demonstrated higher numbers of cytotoxic lymphocytes, B lineage cells, monocyte lineage cells, myeloid dendritic cells, neutrophils, and fibroblasts in the diabetic nephropathy group. The areas under ROC curves for TYROBP, CTSS, LCP2, LUM and TLR7 were 0.9167, 0.9583, 0.9917, 0.93333, and 0.9583, respectively (P < 0.001), and their correlation coefficients with estimated glomerular filtration rate were − 0.8332, − 0.752, − 0.7875, − 0.7567, and − 0.7136, respectively (P < 0.001). The RNA levels of TYROBP, CTSS, LUM and TLR7 were upregulated in high-glucose-treated human renal tubular epithelial cells (P < 0.005). Our study identified TYROBP, CTSS, LCP2, LUM and TLR7 as potentially involved in diabetic nephropathy fibrosis. Furthermore, TYROBP, CTSS, LUM and TLR7 may be associated with epithelial–mesenchymal transition of tubular epithelial cells.







Similar content being viewed by others
Data Availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Alam MS, Sultana A, Sun H, Wu J, Guo F, Li Q, Ren H, Hao Z, Zhang Y, Wang G (2022) Bioinformatics and network-based screening and discovery of potential molecular targets and small molecular drugs for breast cancer. Front Pharmacol 13:942126. https://doi.org/10.3389/fphar.2022.942126
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 4:2. https://doi.org/10.1186/1471-2105-4-2
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) Jvenn: an interactive venn diagram viewer. BMC Bioinform 15:293. https://doi.org/10.1186/1471-2105-15-293
Calle P, Hotter G (2020) Macrophage phenotype and fibrosis in diabetic nephropathy. Int J Mol Sci. https://doi.org/10.3390/ijms21082806
Carew RM, Wang B, Kantharidis P (2012) The role of EMT in renal fibrosis. Cell Tissue Res 347:103–116. https://doi.org/10.1007/s00441-011-1227-1
Cencioni C, Spallotta F, Greco S, Martelli F, Zeiher AM, Gaetano C (2014) Epigenetic mechanisms of hyperglycemic memory. Int J Biochem Cell Biol 51:155–158. https://doi.org/10.1016/j.biocel.2014.04.014
Chevalier RL, Forbes MS, Thornhill BA (2009) Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int 75:1145–1152. https://doi.org/10.1038/ki.2009.86
Colonna M (2003) TREMs in the immune system and beyond. Nat Rev Immunol 3:445–453. https://doi.org/10.1038/nri1106
Deng X, Gao J, Zhao F (2022) Identification of differentially expressed genes and pathways in kidney of ANCA-associated vasculitis by integrated bioinformatics analysis. Ren Fail 44:204–216. https://doi.org/10.1080/0886022X.2022.2030755
Eddy S, Mariani LH, Kretzler M (2020) Integrated multi-omics approaches to improve classification of chronic kidney disease. Nat Rev Nephrol 16:657–668. https://doi.org/10.1038/s41581-020-0286-5
Feng S, Gao Y, Yin D, Lv L, Wen Y, Li Z, Wang B, Wu M, Liu B (2021) Identification of lumican and fibromodulin as hub genes associated with accumulation of extracellular matrix in diabetic nephropathy. Kidney Blood Press Res 46:275–285. https://doi.org/10.1159/000514013
Grayson PC, Eddy S, Taroni JN, Lightfoot YL, Mariani L, Parikh H, Lindenmeyer MT, Ju W, Greene CS, Godfrey B, Cohen CD (2018) Metabolic pathways and immunometabolism in rare kidney diseases. Ann Rheum Dis 77:1226–1233. https://doi.org/10.1136/annrheumdis-2017-212935
Gregg EW, Sattar N, Ali MK (2016) The changing face of diabetes complications. Lancet Diabetes Endocrinol 4:537–547. https://doi.org/10.1016/S2213-8587(16)30010-9
Grigoryev DN, Cheranova DI, Heruth DP et al (2013) Meta-analysis of molecular response of kidney to ischemia reperfusion injury for the identification of new candidate genes. BMC Nephrol 14:231. https://doi.org/10.1186/1471-2369-14-231
Han Y, Wu J, Gong Z, Zhou Y, Li H, Wang B, Qian Q (2021) Identification and development of a novel 5-gene diagnostic model based on immune infiltration analysis of osteoarthritis. J Transl Med 19:522. https://doi.org/10.1186/s12967-021-03183-9
Hanata N, Shoda H, Hatano H et al (2020) Peptidylarginine deiminase 4 promotes the renal infiltration of neutrophils and exacerbates the TLR7 agonist-induced lupus mice. Front Immunol 11:1095. https://doi.org/10.3389/fimmu.2020.01095
Hung PH, Hsu YC, Chen TH, Lin CL (2021) Recent advances in diabetic kidney diseases: from kidney injury to kidney fibrosis. Int J Mol Sci. https://doi.org/10.3390/ijms222111857
Huo Y, Zhang K, Han S, Feng Y, Bao Y (2021) Lymphocyte cytosolic protein 2 is a novel prognostic marker in lung adenocarcinoma. J Int Med Res 49:3000605211059681. https://doi.org/10.1177/03000605211059681
Li Y, Hu Q, Li C et al (2019) PTEN-induced partial epithelial-mesenchymal transition drives diabetic kidney disease. J Clin Invest 129:1129–1151. https://doi.org/10.1172/JCI121987
Li G, Zhang J, Liu D et al (2021) Identification of hub genes and potential ceRNA networks of diabetic nephropathy by weighted gene co-expression network analysis. Front Genet 12:767654. https://doi.org/10.3389/fgene.2021.767654
Lindenmeyer MT, Kretzler M, Boucherot A et al (2007) Interstitial vascular rarefaction and reduced VEGF-A expression in human diabetic nephropathy. J Am Soc Nephrol 18:1765–1776. https://doi.org/10.1681/ASN.2006121304
Liu M, Zen K (2021) Toll-like receptors regulate the development and progression of renal diseases. Kidney Dis (basel) 7:14–23. https://doi.org/10.1159/000511947
Liu J, Kumar S, Dolzhenko E et al (2017) Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight. https://doi.org/10.1172/jci.insight.94716
Loeffler I, Wolf G (2015) Epithelial-to-mesenchymal transition in diabetic nephropathy: fact or fiction. Cells 4:631–652. https://doi.org/10.3390/cells4040631
Lu XM, Ma L, Jin YN, Yu YQ (2015) Lumican overexpression exacerbates lipopolysaccharide-induced renal injury in mice. Mol Med Rep 12:4089–4094. https://doi.org/10.3892/mmr.2015.3940
Lu T, Xu R, Li Q et al (2021) Systematic profiling of ferroptosis gene signatures predicts prognostic factors in esophageal squamous cell carcinoma. Mol Ther Oncolytics 21:134–143. https://doi.org/10.1016/j.omto.2021.02.011
Martínez-Klimova E, Aparicio-Trejo OE, Tapia E, Pedraza-Chaverri J (2019) Unilateral ureteral obstruction as a model to investigate fibrosis-attenuating treatments. Biomolecules. https://doi.org/10.3390/biom9040141
Nastase MV, Zeng-Brouwers J, Wygrecka M, Schaefer L (2018) Targeting renal fibrosis: mechanisms and drug delivery systems. Adv Drug Deliv Rev 129:295–307. https://doi.org/10.1016/j.addr.2017.12.019
Newman AM, Steen CB, Liu CL et al (2019) Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37:773–782. https://doi.org/10.1038/s41587-019-0114-2
Petitprez F, Vano YA, Becht E et al (2018) Transcriptomic analysis of the tumor microenvironment to guide prognosis and immunotherapies. Cancer Immunol Immunother 67:981–988. https://doi.org/10.1007/s00262-017-2058-z
Sommers CL, Menon RK, Grinberg A, Zhang W, Samelson LE, Love PE (2001) Knock-in mutation of the distal four tyrosines of linker for activation of T cells blocks murine T cell development. J Exp Med 194:135–142. https://doi.org/10.1084/jem.194.2.135
Song S, Qiu D, Luo F et al (2018) Knockdown of NLRP3 alleviates high glucose or TGFB1-induced EMT in human renal tubular cells. J Mol Endocrinol 61:101–113. https://doi.org/10.1530/JME-18-0069
Tammaro A, Stroo I, Rampanelli E et al (2013) Role of TREM1-DAP12 in renal inflammation during obstructive nephropathy. PLoS ONE 8:e82498. https://doi.org/10.1371/journal.pone.0082498
Tuleta I, Frangogiannis NG (2021) Diabetic fibrosis. Biochim Biophys Acta Mol Basis Dis 1867:166044. https://doi.org/10.1016/j.bbadis.2020.166044
Wada J, Makino H (2016) Innate immunity in diabetes and diabetic nephropathy. Nat Rev Nephrol 12:13–26. https://doi.org/10.1038/nrneph.2015.175
Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K (2011) Transcriptome analysis of human diabetic kidney disease. Diabetes 60:2354–2369. https://doi.org/10.2337/db10-1181
Wu B, Brooks JD (2012) Gene expression changes induced by unilateral ureteral obstruction in mice. J Urol 188:1033–1041. https://doi.org/10.1016/j.juro.2012.05.004
Xu Z, Jia K, Wang H et al (2021) METTL14-regulated PI3K/Akt signaling pathway via PTEN affects HDAC5-mediated epithelial-mesenchymal transition of renal tubular cells in diabetic kidney disease. Cell Death Dis 12:32. https://doi.org/10.1038/s41419-020-03312-0
Yang J, Dong H, Wang Y et al (2020) Cordyceps cicadae polysaccharides ameliorated renal interstitial fibrosis in diabetic nephropathy rats by repressing inflammation and modulating gut microbiota dysbiosis. Int J Biol Macromol 163:442–456. https://doi.org/10.1016/j.ijbiomac.2020.06.153
Yao X, Cheng F, Yu W et al (2019) Cathepsin S regulates renal fibrosis in mouse models of mild and severe hydronephrosis. Mol Med Rep 20:141–150. https://doi.org/10.3892/mmr.2019.10230
Yayi H, Yeda X, Huaxin W, Yang W, Qian S, Zhongyuan X (2016) Toll-like receptor 7 involves the injury in acute kidney ischemia/reperfusion of STZ-induced diabetic rats. Acta Cir Bras 31:448–455. https://doi.org/10.1590/S0102-865020160070000004
Zeng LF, Xiao Y, Sun L (2019) A glimpse of the mechanisms related to renal fibrosis in diabetic nephropathy. Adv Exp Med Biol 1165:49–79. https://doi.org/10.1007/978-981-13-8871-2_4
Zhang L, Zhang M, Chen X et al (2020) Identification of the tubulointerstitial infiltrating immune cell landscape and immune marker related molecular patterns in lupus nephritis using bioinformatics analysis. Ann Transl Med 8:1596. https://doi.org/10.21037/atm-20-7507
Zheng N, Xie K, Ye H et al (2020) TLR7 in B cells promotes renal inflammation and Gd-IgA1 synthesis in IgA nephropathy. JCI Insight. https://doi.org/10.1172/jci.insight.136965
Zheng W, Guo J, Liu ZS (2021) Effects of metabolic memory on inflammation and fibrosis associated with diabetic kidney disease: an epigenetic perspective. Clin Epigenetics 13:87. https://doi.org/10.1186/s13148-021-01079-5
Zhou LT, Zhang ZJ, Cao JY et al (2021) The unique molecular mechanism of diabetic nephropathy: a bioinformatics analysis of over 250 microarray datasets. Clin Kidney J 14:1626–1638. https://doi.org/10.1093/ckj/sfaa190
Acknowledgements
We thank Wei Chen, PhD, from Beijing Friendship Hospital for his assistance with bioinformatics methodologies. We also thank Jeremy Allen, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing a draft of this manuscript.
Funding
This study was supported by the Wu Jieping Medical Foundation (code 32067502021–11-26) and the Beijing Municipal Administration of Hospitals Incubating Program (code PX2022003).
Author information
Authors and Affiliations
Contributions
The roles and responsibilities of authors were: YB: design, analysis, and interpretation of data, drafting and revising the article. LM: analysis and interpretation of data. DD and DT: providing intellectual content of critical importance to the work described. WL and ZD: providing intellectual content of critical importance to the work described and final approval of the version to be published.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare no conflicts of interest.
Ethical Approval
The data reproduced from the public databases utilized human tissue without identified information. This study was reviewed and deemed exempt by Beijing Friendship Hospital Clinical Research Ethics Committee Review Board.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bai, Y., Ma, L., Deng, D. et al. Title: Bioinformatic Identification of Genes Involved in Diabetic Nephropathy Fibrosis and their Clinical Relevance. Biochem Genet 61, 1567–1584 (2023). https://doi.org/10.1007/s10528-023-10336-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10336-6