Abstract
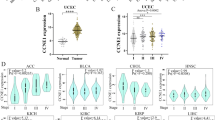
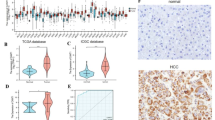
CCDC58, a member of the CCDC protein family, has been primarily associated with the malignant progression of hepatocellular carcinoma (HCC) and breast cancer, with limited research conducted on its involvement in other tumor types. We aimed to assess the significance of CCDC58 in pan-cancer. We utilized the TCGA, GTEx, and UALCAN databases to perform the differential expression of CCDC58 at both mRNA and protein levels. Prognostic value was evaluated through univariate Cox regression and Kaplan–Meier methods. Mutation and methylation analyses were conducted using the cBioPortal and SMART databases. We identified genes interacting with and correlated to CCDC58 through STRING and GEPIA2, respectively. Subsequently, we performed GO and KEGG enrichment analyses. To gain insights into the functional status of CCDC58 at the single-cell level, we utilized CancerSEA. We explored the correlation between CCDC58 and immune infiltration as well as immunotherapy using the ESTIMATE package, TIMER2.0, TISIDB, TIDE, TIMSO, and TCIA. We examined the relationship between CCDC58 and tumor heterogeneity, stemness, DNA methyltransferases, and MMR genes. Lastly, we constructed a nomogram based on CCDC58 in HCC and investigated its association with drug sensitivity. CCDC58 expression was significantly upregulated and correlated with poor prognosis across various tumor types. The mutation frequency of CCDC58 was found to be increased in 25 tumors. We observed a negative correlation between CCDC58 expression and the methylation sites in the majority of tumors. CCDC58 showed negative correlations with immune and stromal scores, as well as with NK T cells, Tregs, CAFs, endothelial cells, and immunomodulators. Its value in immunotherapy was comparable to that of tumor mutational burden. CCDC58 exhibited positive correlations with tumor heterogeneity, stemness, DNA methyltransferase genes, and MMR genes. In HCC, CCDC58 was identified as an independent risk factor and demonstrated potential associations with multiple drugs. CCDC58 demonstrates significant clinical value as a prognostic marker and indicator of immune response across various tumor types. Its comprehensive analysis provides insights into its potential implications in pan-cancer research.








Similar content being viewed by others
Data availability
The data are available from the corresponding author for reasonable requests.
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- AUC:
-
Area under the curve
- BLCA:
-
Bladder urothelial carcinoma
- BP:
-
Biological process
- BRCA:
-
Breast invasive carcinoma
- CAFs:
-
Cancer-associated fibroblasts
- CC:
-
Cellular component
- CCDC58:
-
Coiled-coil domain containing 58
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL:
-
Cholangiocarcinoma
- CNA:
-
Copy number alteration
- CNV:
-
Copy number variants
- COAD:
-
Colon adenocarcinoma
- COADREAD:
-
Colon adenocarcinoma/rectum adenocarcinoma
- DFI:
-
Disease-free interval
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- DSS:
-
Disease-specific survival
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- GBMLGG:
-
Glioma
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- HCC:
-
Hepatocellular carcinoma
- HNSC:
-
Head and neck squamous cell carcinoma
- HRD:
-
Homologous recombination deficiency
- IC50:
-
Half-maximal inhibitory concentration
- ICB:
-
Immune checkpoint blockade
- IPS:
-
Immune phenotype scores
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- KICH:
-
Kidney chromophobe
- KIPAN:
-
Pan-kidney cohort (KICH + KIRC + KIRP)
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- KM:
-
Kaplan–Meier
- LAML:
-
Acute myeloid leukemia
- LGG:
-
Brain lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LOH:
-
Loss of heterozygosity
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MATH:
-
Mutational and clonal intratumoral heterogeneity
- MDSCs:
-
Myeloid-derived suppressor cells
- MESO:
-
Mesothelioma
- MF:
-
Molecular function
- MMR:
-
Mismatch repair
- MSI:
-
Microsatellite instability
- NEO:
-
Neoantigen load
- OS:
-
Overall survival
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PFI:
-
Progression-free interval
- PPI:
-
Protein–protein interaction
- PRAD:
-
Prostate adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- ROC:
-
Receiver operating characteristic
- SARC:
-
Sarcoma
- SKCM:
-
Skin cutaneous melanoma
- SNV:
-
Single nucleotide variants
- STAD:
-
Stomach adenocarcinoma
- STES:
-
Stomach and esophageal carcinoma
- TGCT:
-
Testicular germ cell tumors
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- TILs:
-
Tumor-infiltrating lymphocytes
- TMB:
-
Tumor mutational burden
- TME:
-
Tumor microenvironment
- TNBC:
-
Triple-negative breast cancer
- UCEC:
-
Uterine corpus endometrial carcinoma
- UCS:
-
Uterine carcinosarcoma
- UVM:
-
Uveal melanoma
References
Zoller E, Laborenz J, Kramer L, Boos F, Raschle M, Alexander RT et al (2020) The intermembrane space protein Mix23 is a novel stress-induced mitochondrial import factor. J Biol Chem 295:14686–14697
Liu Z, Yan W, Liu S, Liu Z, Xu P, Fang W (2023) Regulatory network and targeted interventions for CCDC family in tumor pathogenesis. Cancer Lett 565:216225
Pangeni RP, Channathodiyil P, Huen DS, Eagles LW, Johal BK, Pasha D et al (2015) The GALNT9, BNC1 and CCDC8 genes are frequently epigenetically dysregulated in breast tumours that metastasise to the brain. Clin Epigenet 7:57
Deng T, Shen P, Li A, Zhang Z, Yang H, Deng X et al (2021) CCDC65 as a new potential tumor suppressor induced by metformin inhibits activation of AKT1 via ubiquitination of ENO1 in gastric cancer. Theranostics 11:8112–8128
Zhang Z, Xu P, Hu Z, Fu Z, Deng T, Deng X et al (2022) CCDC65, a gene knockout that leads to early death of mice, acts as a potentially novel tumor suppressor in lung adenocarcinoma. Int J Biol Sci 18:4171–4186
Wang Z, Li Y, Yang J, Liang Y, Wang X, Zhang N et al (2022) Circ-TRIO promotes TNBC progression by regulating the miR-432-5p/CCDC58 axis. Cell Death Dis 13:776
Chen L, Zhang J, Yang Y, Shu J, Zheng J, Zhan X et al (2023) Coiled-coil domain-containing protein 58 (CCDC58) is a novel prognostic biomarker correlated with mitochondrial functions in hepatocellular carcinoma. Am J Transl Res 15:2568–2584
Li X, Wang Y, Xu C, Reheman X, Wang Y, Xu R et al (2022) Analysis of competitive endogenous mechanism and survival prognosis of serum exosomes in ovarian cancer patients based on sequencing technology and bioinformatics. Front Genet 13:850089
Kunitomi H, Kobayashi Y, Wu RC, Takeda T, Tominaga E, Banno K et al (2020) LAMC1 is a prognostic factor and a potential therapeutic target in endometrial cancer. J Gynecol Oncol 31:e11
Consortium GT (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45:580–585
Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER et al (2019) Next-generation characterization of the cancer cell line encyclopedia. Nature 569:503–508
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A et al (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38:675–678
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M et al (2022) UALCAN: an update to the integrated cancer data analysis platform. Neoplasia 25:18–27
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A et al (2015) Proteomics: tissue-based map of the human proteome. Science 347:1260419
Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD et al (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173:400-416e11
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H et al (2000) The protein data bank. Nucleic Acids Res 28:235–242
Li Y, Ge D, Lu C (2019) The SMART app: an interactive web application for comprehensive DNA methylation analysis and visualization. Epigenet Chromatin 12:71
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47:W556–W560
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q et al (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48:W509–W514
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2:100141
Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G et al (2019) CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res 47:D900–D908
Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC et al (2019) TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35:4200–4202
Xu L, Deng C, Pang B, Zhang X, Liu W, Liao G et al (2018) TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res 78:6575–6580
Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ et al (2017) Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 1:1–15
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH et al (2018) The immune landscape of cancer. Immunity 48:812–830
Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN et al (2018) Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173:338-354e15
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P et al (2020) Large-scale public data reuse to model immunotherapy response and resistance. Genome Med 12:21
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X et al (2018) Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24:1550–1558
Zeng Z, Wong CJ, Yang L, Ouardaoui N, Li D, Zhang W et al (2022) TISMO: syngeneic mouse tumor database to model tumor immunity and immunotherapy response. Nucleic Acids Res 50:D1391–D1397
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D et al (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18:248–262
Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S et al (2013) Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41:D955–D961
Chi C, Ye Y, Chen B, Huang H (2021) Bipartite graph-based approach for clustering of cell lines by gene expression-drug response associations. Bioinformatics 37:2617–2626
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS et al (2017) Defining a cancer dependency map. Cell 170:564–576
Yang L, Liu Q, Zhang X, Liu X, Zhou B, Chen J et al (2020) DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature 583:133–138
Siriphak S, Chanakankun R, Proungvitaya T, Roytrakul S, Tummanatsakun D, Seubwai W et al (2021) Kallikrein-11, in association with coiled-coil domain containing 25, as a potential prognostic marker for cholangiocarcinoma with lymph node metastasis. Molecules 26:3105
Chanakankun R, Proungvitaya T, Chua-On D, Limpaiboon T, Roytrakul S, Jusakul A et al (2020) Serum coiled-coil domain containing 25 protein as a potential screening/diagnostic biomarker for cholangiocarcinoma. Oncol Lett 19:930–942
Dickson I (2020) NETs promote liver metastasis via CCDC25. Nat Rev Gastroenterol Hepatol 17:451
Gong Y, Qiu W, Ning X, Yang X, Liu L, Wang Z et al (2015) CCDC34 is up-regulated in bladder cancer and regulates bladder cancer cell proliferation, apoptosis and migration. Oncotarget 6:25856–25867
Geng W, Liang W, Fan Y, Ye Z, Zhang L (2018) Overexpression of CCDC34 in colorectal cancer and its involvement in tumor growth, apoptosis and invasion. Mol Med Rep 17:465–473
Zhou M, Chen X, Bai H, Sun Y, Zhang Z, Li S et al (2021) RABL2A-CCDC34 axis promotes sorafenib resistance in hepatocellular carcinoma. DNA Cell Biol 40:1418–1427
Liu LB, Huang J, Zhong JP, Ye GL, Xue L, Zhou MH et al (2018) High expression of CCDC34 is associated with poor survival in cervical cancer patients. Med Sci Monit 24:8383–8390
Wang J, Wu X, Dai W, Li J, Xiang L, Tang W et al (2020) The CCDC43-ADRM1 axis regulated by YY1, promotes proliferation and metastasis of gastric cancer. Cancer Lett 482:90–101
Chen Y, Pei M, Li J, Wang Z, Liu S, Xiang L et al (2022) Disruption of the CCDC43-FHL1 interaction triggers apoptosis in gastric cancer cells. Exp Cell Res 415:113107
Wang J, Liu G, Liu M, Xiang L, Xiao Y, Zhu H et al (2018) The FOXK1-CCDC43 axis promotes the invasion and metastasis of colorectal cancer cells. Cell Physiol Biochem 51:2547–2563
Lin H, Gao Y, Sun K, Zhang Q, Li Y, Chen M et al (2022) COA3 overexpression promotes non-small cell lung cancer metastasis by reprogramming glucose metabolism. Am J Cancer Res 12:3662–3678
Xu X, Cao W, Sun W, Wang Z, Chen H, Zheng Z et al (2019) Knockdown of CCDC132 attenuates gastric cancer cells proliferation and tumorigenesis by facilitating DNA damage signaling. Cancer Manag Res 11:9585–9597
Chen PS, Hsu HP, Phan NN, Yen MC, Chen FW, Liu YW et al (2021) CCDC167 as a potential therapeutic target and regulator of cell cycle-related networks in breast cancer. Aging 13:4157–4181
Hu X, Zhao Y, Wei L, Zhu B, Song D, Wang J et al (2017) CCDC178 promotes hepatocellular carcinoma metastasis through modulation of anoikis. Oncogene 36:4047–4059
Bagci O, Kurtgoz S (2015) Amplification of Cellular oncogenes in solid tumors. N Am J Med Sci 7:341–346
Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM (2015) Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 149:1226-1239e4
Long J, Wang A, Bai Y, Lin J, Yang X, Wang D et al (2019) Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine 42:363–374
Cancer Genome Atlas Research Network (2017) Electronic address wbe, cancer genome atlas research N. comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169:1327–1341
Ramalho-Carvalho J, Henrique R, Jeronimo C (2018) Methylation-specific PCR. Methods Mol Biol 1708:447–472
Kaaij LJ, Mokry M, Zhou M, Musheev M, Geeven G, Melquiond AS et al (2016) Enhancers reside in a unique epigenetic environment during early zebrafish development. Genome Biol 17:146
Shapovalov Y, Hoffman D, Zuch D, de Mesy Bentley KL, Eliseev RA (2011) Mitochondrial dysfunction in cancer cells due to aberrant mitochondrial replication. J Biol Chem 286:22331–22338
Yang Y, Pan C, Yu L, Ruan H, Chang L, Yang J et al (2019) SSBP1 upregulation in colorectal cancer regulates mitochondrial mass. Cancer Manag Res 11:10093–10106
Jiang HL, Sun HF, Gao SP, Li LD, Huang S, Hu X et al (2016) SSBP1 suppresses TGFbeta-driven epithelial-to-mesenchymal transition and metastasis in triple-negative breast cancer by regulating mitochondrial retrograde signaling. Cancer Res 76:952–964
Su J, Li Y, Liu Q, Peng G, Qin C, Li Y (2022) Identification of SSBP1 as a ferroptosis-related biomarker of glioblastoma based on a novel mitochondria-related gene risk model and in vitro experiments. J Transl Med 20:440
Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ (2011) Natural innate and adaptive immunity to cancer. Annu Rev Immunol 29:235–271
Hallett WH, Murphy WJ (2004) Natural killer cells: biology and clinical use in cancer therapy. Cell Mol Immunol 1:12–21
Wang K, Yu M, Zhang Z, Yin R, Chen Q, Zhao X et al (2023) Integrated analysis of single-cell and bulk transcriptome identifies a signature based on NK cell marker genes to predict prognosis and therapeutic response in clear cell renal cell carcinoma. Transl Cancer Res 12:1270–1289
Na HY, Park Y, Nam SK, Koh J, Kwak Y, Ahn SH et al (2021) Prognostic significance of natural killer cell-associated markers in gastric cancer: quantitative analysis using multiplex immunohistochemistry. J Transl Med 19:529
Wang F, Lau JKC, Yu J (2021) The role of natural killer cell in gastrointestinal cancer: killer or helper. Oncogene 40:717–730
Reid FSW, Egoroff N, Pockney PG, Smith SR (2021) A systematic scoping review on natural killer cell function in colorectal cancer. Cancer Immunol Immunother 70:597–606
Diaz-Montero CM, Rini BI, Finke JH (2020) The immunology of renal cell carcinoma. Nat Rev Nephrol 16:721–735
Ping G, Tian Y, Zhou Z (2022) Constructing a tregs-associated signature to predict the prognosis of colorectal cancer patients: a STROBE-compliant retrospective study. Medicine 101:e31382
Zimmer N, Trzeciak ER, Graefen B, Satoh K, Tuettenberg A (2022) GARP as a therapeutic target for the modulation of regulatory T cells in cancer and autoimmunity. Front Immunol 13:928450
Shang B, Liu Y, Jiang SJ, Liu Y (2015) Prognostic value of tumor-infiltrating FoxP3 + regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep 5:15179
Winerdal ME, Marits P, Winerdal M, Hasan M, Rosenblatt R, Tolf A et al (2011) FOXP3 and survival in urinary bladder cancer. BJU Int 108:1672–1678
Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM et al (2020) A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 20:174–186
Kalluri R (2016) The biology and function of fibroblasts in cancer. Nat Rev Cancer 16:582–598
Kalluri R, Zeisberg M (2006) Fibroblasts in cancer. Nat Rev Cancer 6:392–401
Maishi N, Hida K (2017) Tumor endothelial cells accelerate tumor metastasis. Cancer Sci 108:1921–1926
Sobierajska K, Ciszewski WM, Sacewicz-Hofman I, Niewiarowska J (2020) Endothelial cells in the tumor microenvironment. Adv Exp Med Biol 1234:71–86
Phuengkham H, Ren L, Shin IW, Lim YT (2019) Nanoengineered immune niches for reprogramming the immunosuppressive tumor microenvironment and enhancing cancer immunotherapy. Adv Mater 31:e1803322
Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A et al (2019) Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 30:44–56
Yamamoto H, Imai K (2015) Microsatellite instability: an update. Arch Toxicol 89:899–921
Toh M, Ngeow J (2021) Homologous recombination deficiency: cancer predispositions and treatment implications. Oncologist 26:e1526–e1537
Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A et al (2019) Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci USA 116:9020–9029
Saygin C, Matei D, Majeti R, Reizes O, Lathia JD (2019) Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell 24:25–40
Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D et al (2011) The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8:511–524
Siu MK, Wong ES, Kong DS, Chan HY, Jiang L, Wong OG et al (2013) Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene 32:3500–3509
Yin ZQ, Liu JJ, Xu YC, Yu J, Ding GH, Yang F et al (2014) A 41-gene signature derived from breast cancer stem cells as a predictor of survival. J Exp Clin Cancer Res 33:49
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
HW, QG, WS, and CQ participated in the conception, design, acquisition of data, analysis, and interpretation. All authors participated in drafting the article and gave final approval.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, H., Geng, Q., Shi, W. et al. Comprehensive pan-cancer analysis reveals CCDC58 as a carcinogenic factor related to immune infiltration. Apoptosis 29, 536–555 (2024). https://doi.org/10.1007/s10495-023-01919-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-023-01919-0