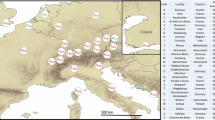
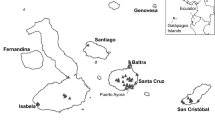
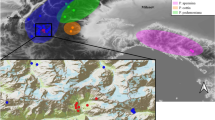
Abstract
Native to Southeastern Asia, the ambrosia beetle Xylosandrus compactus is invasive worldwide. Its invasion is favoured by its cryptic lifestyle, symbiosis with a fungus that facilitates a broad range of host plants, and predominant sib-mating reproduction. X. compactus invaded Africa more than a century ago and the Americas and Pacific Islands in the middle of the twentieth century. It was not detected in Europe before 2011, when it was first reported in Italy before quickly spreading to France, Greece and Spain. Despite the negative environmental, agricultural and economic consequences of the invasion of X. compactus, its invasion history and main pathways remain poorly documented. We used COI and RAD sequencing to (i) characterise the worldwide genetic structure of the species, (ii) disentangle the origin(s) of the non-native populations on the three invaded continents and (iii) analyse the genetic diversity and pathways within each invaded region. Three mitochondrial lineages were identified in the native range. Populations invading Europe and the American-Pacific region originated from the first lineage and were only slightly genetically differentiated at nuclear SNP markers, suggesting independent introductions from close sources in or near Shanghai, ca. 60 years apart. Populations invading Africa originated from the second lineage, likely from India or Vietnam.


Similar content being viewed by others
Availability of data and material
Individual RAD sequence files are available in a fq.gz format at the Sequence Read Archive (SRA) (Study Accession no. PRJNA771401). The VCF files Xylosandruscompactus.vcf and Ambrosiellaxylebori.vcf used for population genomic analyses, as well as the popmap used in STACK’ population module and specimens' metadata (e.g., GPS coordinates) are available on Portail Data INRAE (https://doi.org/10.15454/ETBWEP). The Genbank accession numbers for the mitochondrial haplotypes reported in this paper are OK489329:OK489334.
References
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19(9):1655–1664. https://doi.org/10.1101/gr.094052.109
Andersen HF, Jordal BH, Kambestad M, Kirkendall LR (2012) Improbable but true: the invasive inbreeding ambrosia beetle Xylosandrus morigerus has generalist genotypes. Ecol Evol 2(1):247–257. https://doi.org/10.1002/ece3.58
Andreev D, Breilid H, Kirkendall L, Brun LO, Ffrench-Constant RH (1998) Lack of nucleotide variability in a beetle pest with extreme inbreeding. Insect Mol Biol 7(2):197–200. https://doi.org/10.1046/j.1365-2583.1998.72064.x
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3(10):e3376. https://doi.org/10.1371/journal.pone.0003376
Baker HG (1965) Characteristics and modes of origin in weeds. In: Baker HG SG, editors (ed) The Genetics of Colonizing Species. Academic Press, New York, NY, USA, pp 147–172
Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16(1):37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Beaver RA, Sittichaya W, Liu LY (2014) A synopsis of the scolytine ambrosia beetles of Thailand (Coleoptera: Curculionidae: Scolytinae). Zootaxa 3875:1–82. https://doi.org/10.11646/zootaxa.3875.1.1
Becker RA, Wilks AR, Brownrigg R, Minka TP, Deckmyn A (2018) maps: draw geographical maps. R package version 3.3.0.
Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S (2016) pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32(18):2817–2823. https://doi.org/10.1093/bioinformatics/btw327
Bosso L, Senatore M, Varlese R, Ruocco M, Garonna AP, Bonanomi G, Mazzoleni S, Cristinzio G (2012) Severe outbreak of Fusarium solani on Quercus ilex vectored by Xylosandrus compactus. J Plant Pathol 94:85–105. https://doi.org/10.4454/JPP.V95I4SUP.036
Bradshaw CJ, Leroy B, Bellard C, Roiz D, Albert C, Fournier A, Barbet-Massin M, Salles JM, Simard F, Courchamp F (2016) Massive yet grossly underestimated global costs of invasive insects. Nat Commun 7:12986. https://doi.org/10.1038/ncomms12986
Bras A, Avtzis DN, Kenis M, Li H, Vétek G, Bernard A, Courtin C, Rousselet J, Roques A, Auger-Rozenberg M-A (2019) A complex invasion story underlies the fast spread of the invasive box tree moth (Cydalima perspectalis) across Europe. J Pest Sci 92(3):1187–1202. https://doi.org/10.1007/s10340-019-01111-x
Browne FG (1961) The biology of Malayan Scolytidae and Platypodidae. Malay for Rec 22:1–255
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22(11):3124–3140. https://doi.org/10.1111/mec.12354
Cognato AI, Sari G, Smith SM, Beaver RA, Li Y, Hulcr J, Jordal BH, Kajimura H, Lin C-S, Pham TH, Singh S, Sittichaya W (2020) The essential role of taxonomic expertise in the creation of DNA databases for the identification and delimitation of Southeast Asian ambrosia beetle species (Curculionidae: Scolytinae: Xyleborini). Front Ecol Evol. https://doi.org/10.3389/fevo.2020.00027
Cruaud A, Gautier M, Rossi JP, Rasplus JY, Gouzy J (2016) RADIS: analysis of RAD-seq data for interspecific phylogeny. Bioinformatics 32(19):3027–3028. https://doi.org/10.1093/bioinformatics/btw352
Cruaud A, Groussier G, Genson G, Saune L, Polaszek A, Rasplus JY (2018) Pushing the limits of whole genome amplification: successful sequencing of RADseq library from a single microhymenopteran (Chalcidoidea, Trichogramma). PeerJ 6:e5640. https://doi.org/10.7717/peerj.5640
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, De Pristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, Genomes Project Analysis G (2011) The variant call format and VCFtools. Bioinformatics 27 (15):2156-2158. https://doi.org/10.1093/bioinformatics/btr330
Davey JW, Blaxter ML (2010) RADSeq: next-generation population genetics. Brief Funct Genom 9(5–6):416–423. https://doi.org/10.1093/bfgp/elq031
Dole SA, Jordal BH, Cognato AI (2010) Polyphyly of Xylosandrus Reitter inferred from nuclear and mitochondrial genes (Coleoptera: Curculionidae: Scolytinae). Mol Phylogen Evol 54(3):773–782. https://doi.org/10.1016/j.ympev.2009.11.011
Dueñas M-A, Hemming DJ, Roberts A, Diaz-Soltero H (2021) The threat of invasive species to IUCN-listed critically endangered species: A systematic review. Glob Ecol Conserv. https://doi.org/10.1016/j.gecco.2021.e01476
Dzurenko M, Ranger CM, Hulcr J, Galko J, Kaňuch P (2020) Origin of non-native Xylosandrus germanus, an invasive pest ambrosia beetle in Europe and North America. J Pest Sci 94(2):553–562. https://doi.org/10.1007/s10340-020-01283-x
Egonyu JP, Baguma J, Ogari I, Ahumuza G, Kyamanywa S, Kucel P, Kagezi GH, Erbaugh M, Phiri N, Ritchie BJ, Wagoire WW (2015) The formicid ant, Plagiolepis sp., as a predator of the coffee twig borer Xylosandrus Compactus. Biol Control 91:42–46. https://doi.org/10.1016/j.biocontrol.2015.07.011
Estoup A, Guillemaud T (2010) Reconstructing routes of invasion using genetic data: why, how and so what? Mol Ecol 19(19):4113–4130. https://doi.org/10.1111/j.1365-294X.2010.04773.x
Estoup A, Ravigné V, Hufbauer R, Vitalis R, Gautier M, Facon B (2016) Is there a genetic paradox of biological invasion? Annu Rev Ecol Evol Syst 47(1):51–72. https://doi.org/10.1146/annurev-ecolsys-121415-032116
Etter PD, Bassham S, Hohenlohe PA, Johnson EA, Cresko WA (2011) SNP discovery and genotyping for evolutionary genetics using RAD sequencing. Methods Mol Biol 772:157–178. https://doi.org/10.1007/978-1-61779-228-1_9
Evans EA, Crane J, Hodges A, Osborne JL (2010) Potential economic impact of laurel wilt disease on the Florida avocado industry. HortTechnology 20(1):234–238. https://doi.org/10.21273/horttech.20.1.234
Eyer PA, Matsuura K, Vargo EL, Kobayashi K, Yashiro T, Suehiro W, Himuro C, Yokoi T, Guenard B, Dunn RR, Tsuji K (2018) Inbreeding tolerance as a pre-adapted trait for invasion success in the invasive ant Brachyponera chinensis. Mol Ecol 27(23):4711–4724. https://doi.org/10.1111/mec.14910
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3(5):294–299
Francis RM (2017) pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour 17(1):27–32. https://doi.org/10.1111/1755-0998.12509
Gallego D, María Riba J, Molina N, González E, di Sora N, Núñez L, María Closa A, Comparini C, Leza M (2020) Las invasiones silenciosas de escolítidos: el caso del género Xylosandrus (Coleoptera, Curculionidae, Scolytinae). Foresta 78:78–83
Gao L, Wu S, Lu G, Chi X, Ji G, Zhu Y (2017) Morphological characteristics and damage survey of Xylosandrus compactus. For Pest Dis 36:31–34
Garonna AP, Dole SA, Saracino A, Mazzoleni S, Cristinzio G (2012) First record of the black twig borer Xylosandrus compactus (Eichhoff) (Coleoptera: Curculionidae, Scolytinae) from Europe. Zootaxa 3251(3251):64–68. https://doi.org/10.11646/zootaxa.3251.1.5
Gascoigne J, Berec L, Gregory S, Courchamp F (2009) Dangerously few liaisons: a review of mate-finding Allee effects. Popul Ecol 51(3):355–372. https://doi.org/10.1007/s10144-009-0146-4
Gippet JMW, Liebhold AM, Fenn-Moltu G, Bertelsmeier C (2019) Human-mediated dispersal in insects. Curr Opin Insect Sci 35:96–102. https://doi.org/10.1016/j.cois.2019.07.005
Guangchuang Y (2020) scatterpie: Scatter Pie Plot. R package version 0.1.5.
Gugliuzzo A, Criscione G, Siscaro G, Russo A, Tropea Garzia G (2019) First data on the flight activity and distribution of the ambrosia beetle Xylosandrus compactus (Eichhoff) on carob trees in Sicily. EPPO Bulletin 49(2):340–351. https://doi.org/10.1111/epp.12564
Gugliuzzo A, Biedermann PHW, Carrillo D, Castrillo LA, Egonyu JP, Gallego D, Haddi K, Hulcr J, Jactel H, Kajimura H, Kamata N, Meurisse N, Li Y, Oliver JB, Ranger CM, Rassati D, Stelinski LL, Sutherland R, Tropea Garzia G, Wright MG, Biondi A (2021) Recent advances toward the sustainable management of invasive Xylosandrus ambrosia beetles. J Pest Sci 94(3):615–637. https://doi.org/10.1007/s10340-021-01382-3
Hara AH, Beardsley JW Jr (1979) The biology of the black twig borer, Xylosandrus compactus (Eichhoff). Hawaii Proc Hawaii Entomol Soc 13(1):55–70
Hulcr J, Dunn RR (2011) The sudden emergence of pathogenicity in insect-fungus symbioses threatens naive forest ecosystems. Proc R Soc b: Biol Sci 278(1720):2866–2873. https://doi.org/10.1098/rspb.2011.1130
Hulcr J, Black A, Prior K, Chen C-Y, Li H-F (2017) Studies of ambrosia beetles (Coleoptera: Curculionidae) in their native ranges help predict invasion impact. Fla Entomol 100(2):257–261. https://doi.org/10.1653/024.100.0219
Ito M, Kajimura H (2009) Phylogeography of an ambrosia beetle, Xylosandrus crassiusculus (Motschulsky) (Coleoptera: Curculionidae: Scolytinae). Japan Appl Entomol Zool 44(4):549–559. https://doi.org/10.1303/aez.2009.549
Jones BA (2017) Invasive species impacts on human well-being using the life satisfaction index. Ecol Econ 134:250–257. https://doi.org/10.1016/j.ecolecon.2017.01.002
Jones BA, McDermott SM (2017) Health impacts of invasive species through an altered natural environment: assessing air pollution sinks as a causal pathway. Environ Resour Econ 71(1):23–43. https://doi.org/10.1007/s10640-017-0135-6
Jordal BH, Beaver RA, Kirkendall LR (2001) Breaking taboos in the tropics: incest promotes colonization by wood-boring beetles. Global Ecol Biogeogr 10(4):345–357. https://doi.org/10.1046/j.1466-822X.2001.00242.x
Kenis M, Auger-Rozenberg M-A, Roques A, Timms L, Péré C, Cock MJW, Settele J, Augustin S, Lopez-Vaamonde C (2008) Ecological effects of invasive alien insects. Biol Invasions 11(1):21–45. https://doi.org/10.1007/s10530-008-9318-y
Kiran VS, Asokan R, Revannavar R, Hanchipura Mallesh MS, Ramasamy E (2019) Genetic characterization and DNA barcoding of coffee shot-hole borer, Xylosandrus compactus (Eichhoff) (Coleoptera: Curculionidae: Scolytinae). Mitochondrial DNA Part A 30(7):779–785. https://doi.org/10.1080/24701394.2019.1659249
Kirkendall LR, Odegaard F (2007) Ongoing invasions of old-growth tropical forests: establishment of three incestuous beetle species in southern Central America (Curculionidae : Scolytinae). Zootaxa 1588:53–62. https://doi.org/10.11646/zootaxa.1588.1.3
Kirkendall LR, Dal Cortivo M, Gatti E (2008) First record of the ambrosia beetle, Monarthrum mali (Curculionidae, Scolytinae) in Europe. J Pest Sci 81(3):175–178. https://doi.org/10.1007/s10340-008-0196-y
Kirkendall LR, Biedermann PHW, Jordal BH (2015) Evolution and diversity of bark and ambrosia beetles. In: Bark Beetles. pp 85–156. https://doi.org/10.1016/b978-0-12-417156-5.00003-4
Kühnholz S, Borden JH, Uzunovic A (2001) Secondary ambrosia beetles in apparently healthy trees: adaptations, potential causes and suggested research. Integr Pest Manag Rev 6(3/4):209–219. https://doi.org/10.1023/a:1025702930580
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
LaBonte JR (2010) Eradictation of an exotic ambrosia beetle, Xylosandrus crassiusculus (Motschulsky), in Oregon. In: USDA Research Forum on Invasive Species, pp 41–43
Landi L, Gómez D, Braccini CL, Pereyra VA, Smith SM, Marvaldi AE (2017) Morphological and molecular identification of the invasive Xylosandrus crassiusculus (Coleoptera: Curculionidae: Scolytinae) and its South American range extending into Argentina and Uruguay. Ann Entomol Soc Am 110(3):344–349. https://doi.org/10.1093/aesa/sax032
Leza MAR, Nuñez L, Riba JM, Comparini C, Roca Á, Gallego D (2020) First record of the black twig borer, Xylosandrus compactus (Coleoptera: Curculionidae, Scolytinae) in Spain. Zootaxa 4767(2):345–350. https://doi.org/10.11646/zootaxa.4767.2.9
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Linck E, Battey CJ (2019) Minor allele frequency thresholds strongly affect population structure inference with genomic data sets. Mol Ecol Resour 19(3):639–647. https://doi.org/10.1111/1755-0998.12995
Liu M, Zhang D, Pietzarka U, Roloff A (2021) Assessing the adaptability of urban tree species to climate change impacts: a case study in Shanghai. Urban for Urban Green. https://doi.org/10.1016/j.ufug.2021.127186
Lombaert E, Guillemaud T, Cornuet JM, Malausa T, Facon B, Estoup A (2010) Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS ONE 5(3):e9743. https://doi.org/10.1371/journal.pone.0009743
Mitchell A, Maddox C (2010) Bark beetles (Coleoptera: Curculionidae: Scolytinae) of importance to the Australian macadamia industry: an integrative taxonomic approach to species diagnostics. Aust J Entomol 49(2):104–113. https://doi.org/10.1111/j.1440-6055.2010.00746.x
Morales CL, Saez A, Garibaldi LA, Aizen MA (2017) Disruption of pollination services by invasive pollinator species, vol. 12. Impact of Biological Invasions on Ecosystem Services https://doi.org/10.1007/978-3-319-45121-3_13
Nei M (1972) Genetic distance between populations. Am Nat 106(949):283–292. https://doi.org/10.1086/282771
Ngoan ND, Wilkinson RC, Short DE, Moses CS, Mangold JR (1976) Biology of an introduced ambrosia beetle, Xylosandrus compactus. Florida Ann Entomol Soc Am 69(5):872–876. https://doi.org/10.1093/aesa/69.5.872
Oliveira CM, Flechtmann CAH, Frizzas MR (2008) First record of Xylosandrus compactus (Eichhoff) (Coleoptera: Curculionidae: Scolytinae) on soursop, Annona muricata L. (Annonaceae) in Brazil, with a list of host plants. Coleopt Bull 62(1):45–48. https://doi.org/10.1649/1039.1
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. R package version 0.14. Bioinformatics 26:419–420
Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A (2010) How many bootstrap replicates are necessary? J Comput Biol 17(3):337–354. https://doi.org/10.1089/cmb.2009.0179
Peer K, Taborsky M (2005) Outbreeding depression, but no inbreeding depression in haplodiploid ambrosia beetles with regular sibling mating. Evolution 59(2):317–323. https://doi.org/10.1111/j.0014-3820.2005.tb00992.x
Pejchar L, Mooney HA (2009) Invasive species, ecosystem services and human well-being. Trends Ecol Evol 24(9):497–504. https://doi.org/10.1016/j.tree.2009.03.016
Pembleton LW, Cogan NO, Forster JW (2013) StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol Ecol Resour 13(5):946–952. https://doi.org/10.1111/1755-0998.12129
Ploetz RC, Hulcr J, Wingfield MJ, de Beer ZW (2013) Destructive tree diseases associated with ambrosia and bark beetles: black swan events in tree pathology? Plant Dis 97(7):856–872. https://doi.org/10.1094/PDIS-01-13-0056-FE
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
Raffa KF, Grégoire J-C, Staffan Lindgren B (2015) Natural history and ecology of bark beetles. In: Vega FE, Hofstetter RW (eds) Bark Beetles. Biology and Ecology of Native and Invasive Species. Elsevier, pp 585–614. https://doi.org/10.1016/b978-0-12-417156-5.00001-0
Ranger CM, Schultz PB, Frank SD, Chong JH, Reding ME (2015) Non-native ambrosia beetles as opportunistic exploiters of living but weakened trees. PLoS ONE 10(7):e0131496. https://doi.org/10.1371/journal.pone.0131496
Rizzo D, Da Lio D, Bartolini L, Salemi C, Del Nista D, Aronadio A, Pennacchio F, Binazzi F, Francardi V, Garonna AP, Rossi E (2021) TaqMan probe assays on different biological samples for the identification of three ambrosia beetle species, Xylosandrus compactus (Eichoff), X. crassiusculus (Motschulsky) and X. germanus (Blandford) (Coleoptera Curculionidae Scolytinae). 3 Biotech 11 (6):259. https://doi.org/10.1007/s13205-021-02786-9
Rochette NC, Rivera-Colon AG, Catchen JM (2019) Stacks 2: analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol Ecol 28(21):4737–4754. https://doi.org/10.1111/mec.15253
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542. https://doi.org/10.1093/sysbio/sys029
Roques A, Bellanger R, Daubrée JB, Ducatillon C, Urvois T, Auger-Rozenberg MA (2019) Les scolytes exotiques : une menace pour le maquis. Phytoma 727:16–20
Sardain A, Sardain E, Leung B (2019) Global forecasts of shipping traffic and biological invasions to 2050. Nat Sustain 2(4):274–282. https://doi.org/10.1038/s41893-019-0245-y
Sax DF, Brown JH (2000) The paradox of invasion. Global Ecol Biogeogr 9(5):363–371. https://doi.org/10.1046/j.1365-2699.2000.00217.x
Schindler S, Staska B, Adam M, Rabitsch W, Essl F (2015) Alien species and public health impacts in Europe: a literature review. Neobiota 27:1–23. https://doi.org/10.3897/neobiota.27.5007
Schrieber K, Lachmuth S (2017) The genetic paradox of invasions revisited: the potential role of inbreeding x environment interactions in invasion success. Biol Rev Camb Philos Soc 92(2):939–952. https://doi.org/10.1111/brv.12263
Seebens H, Blackburn TM, Dyer EE, Genovesi P, Hulme PE, Jeschke JM, Pagad S, Pysek P, Winter M, Arianoutsou M, Bacher S, Blasius B, Brundu G, Capinha C, Celesti-Grapow L, Dawson W, Dullinger S, Fuentes N, Jager H, Kartesz J, Kenis M, Kreft H, Kuhn I, Lenzner B, Liebhold A, Mosena A, Moser D, Nishino M, Pearman D, Pergl J, Rabitsch W, Rojas-Sandoval J, Roques A, Rorke S, Rossinelli S, Roy HE, Scalera R, Schindler S, Stajerova K, Tokarska-Guzik B, van Kleunen M, Walker K, Weigelt P, Yamanaka T, Essl F (2017) No saturation in the accumulation of alien species worldwide. Nat Commun 8:14435. https://doi.org/10.1038/ncomms14435
Seebens H, Bacher S, Blackburn TM, Capinha C, Dawson W, Dullinger S, Genovesi P, Hulme PE, van Kleunen M, Kuhn I, Jeschke JM, Lenzner B, Liebhold AM, Pattison Z, Pergl J, Pysek P, Winter M, Essl F (2020) Projecting the continental accumulation of alien species through to 2050. Global Change Biol. https://doi.org/10.1111/gcb.15333
Simberloff D, Martin JL, Genovesi P, Maris V, Wardle DA, Aronson J, Courchamp F, Galil B, Garcia-Berthou E, Pascal M, Pysek P, Sousa R, Tabacchi E, Vila M (2013) Impacts of biological invasions: what’s what and the way forward. Trends Ecol Evol 28(1):58–66. https://doi.org/10.1016/j.tree.2012.07.013
Spanou K, Marathianou M, Gouma M, Dimou D, Nikoletos L, Milonas P, Papachristos D (2019) First record of black twig borer Xylosandrus compactus (Coleoptera: Curculionidae) in Greece. Paper presented at the 18th Panhellenic Entomological Congress, Komotini, 15–17/10/2019
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Storer C, Payton A, McDaniel S, Jordal B, Hulcr J (2017) Cryptic genetic variation in an inbreeding and cosmopolitan pest, Xylosandrus crassiusculus, revealed using ddRADseq. Ecol Evol 7(24):10974–10986. https://doi.org/10.1002/ece3.3625
Stouthamer R, Rugman-Jones P, Thu PQ, Eskalen A, Thibault T, Hulcr J, Wang L-J, Jordal BH, Chen C-Y, Cooperband M, Lin C-S, Kamata N, Lu S-S, Masuya H, Mendel Z, Rabaglia R, Sanguansub S, Shih H-H, Sittichaya W, Zong S (2017) Tracing the origin of a cryptic invader: phylogeography of the Euwallacea fornicatus (Coleoptera: Curculionidae: Scolytinae) species complex. Agric for Entomol 19(4):366–375. https://doi.org/10.1111/afe.12215
Toews DPL, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21(16):3907–3930. https://doi.org/10.1111/j.1365-294X.2012.05664.x
UNCTAD (2020) Review of maritime transport 2020. United Nations, Geneva
Urvois T, Auger-Rozenberg MA, Roques A, Rossi JP, Kerdelhue C (2021) Climate change impact on the potential geographical distribution of two invading Xylosandrus ambrosia beetles. Sci Rep 11(1):1339. https://doi.org/10.1038/s41598-020-80157-9
Vanderpool D, Bracewell RR, McCutcheon JP (2018) Know your farmer: ancient origins and multiple independent domestications of ambrosia beetle fungal cultivars. Mol Ecol 27(8):2077–2094. https://doi.org/10.1111/mec.14394
Wang M, Li J, Kuang S, He Y, Chen G, Huang Y, Song C, Anderson P, Łowicki D (2020) Plant diversity along the urban-rural gradient and its relationship with urbanization degree in Shanghai, China. Forests https://doi.org/10.3390/f11020171
Wasser SK, Shedlock AM, Comstock K, Ostrander EA, Mutayoba B, Stephens M (2004) Assigning African elephant DNA to geographic region of origin: applications to the ivory trade. Proc Natl Acad Sci USA 101(41):14847–14852. https://doi.org/10.1073/pnas.0403170101
Weber BC, McPherson JE (1983) World list of host plants of Xylosandrus germanus (Blandford) (Coleoptera: Scolytidae). Coleopt Bull 37(2):114–134
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. https://doi.org/10.2307/2408641
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York
Wood SL (1980) New American bark beetles (Coleoptera: Scolytidae) with two recently introduced species. Great Basin Nat 40(4):353–358
Wright S (1951) The genetical structure of populations. Ann Eugen 15(4):323–354. https://doi.org/10.1111/j.1469-1809.1949.tb02451.x
Zheng X (2013) Statistical prediction of HLA alleles and relatedness analysis in genome-wide association studies. University of Washington, Washington
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24):3326–3328. https://doi.org/10.1093/bioinformatics/bts606
Acknowledgements
We would like to thank Richard Bellanger, Jared Bernard, Jean-Baptiste Daubrée, Jacques Deleuze, Massimo Faccoli, Steve Frank, Lei Gao (Shanghai Academy of Landscape Architecture Science and Planning), Benjamin Gey, Federica Giarruzzo, Fabrice Pinard, and Giovanna Tropea for sending specimens. We also would like to thank Anne Loiseau for the laboratory work. We are grateful to MGX-Montpellier GenomiX for sequencing the RAD libraries and to the Genotoul bioinformatics platform Toulouse Occitanie (Bioinfo Genotoul, https://doi.org/10.15454/1.5572369328961167E12) for providing computing and storage resources. We are also grateful to the Sino-French Joint Laboratory for Invasive Forest Pests in Eurasia (IFOPE) for its support.
Funding
This work was supported by the LIFE project SAMFIX (SAving Mediterranean Forests from Invasions of Xylosandrus beetles and associated pathogenic fungi, LIFE17 NAT/IT/000609, https://www.lifesamfix.eu/) which received funding from the European Union’s LIFE Nature and Biodiversity programme. JH and AJJ were supported by the US Forest Service, the USDA APHIS, and the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests. Specimens sampled did not involve endangered nor protected species.
Additional information
Communicated by Antonio Biondi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Urvois, T., Perrier, C., Roques, A. et al. A first inference of the phylogeography of the worldwide invader Xylosandrus compactus. J Pest Sci 95, 1217–1231 (2022). https://doi.org/10.1007/s10340-021-01443-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-021-01443-7