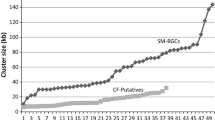
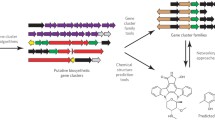
Abstract
Natural products are a crucial source of antimicrobial agents, but reliance on low-resolution bioactivity-guided approaches has led to diminishing interest in discovery programmes. Here, we demonstrate that two in-house automated informatic platforms can be used to target classes of biologically active natural products, specifically, peptaibols. We demonstrate that mass spectrometry-based informatic approaches can be used to detect natural products with high sensitivity, identifying desired agents present in complex microbial extracts. Using our specialised software packages, we could elaborate specific branches of chemical space, uncovering new variants of trichopolyn and demonstrating a way forward in mining natural products as a valuable source of potential pharmaceutical agents.



Similar content being viewed by others
References
Bachmann BO, Van Lanen SG, Baltz RH (2014) Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol 41(2):175–184. doi:10.1007/s10295-013-1389-9
Backman TW, Cao Y, Girke T (2011) ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res 39(Web Server issue):W486W–491. doi:10.1093/nar/gkr320
Chugh JK, Wallace BA (2001) Peptaibols: models for ion channels. Biochem Soc Trans 29(Pt 4):565–570
Clardy J, Fischbach MA, Walsh CT (2006) New antibiotics from bacterial natural products. Nat Biotechnol 24(12):1541–1550. doi:10.1038/nbt1266
Degenkolb T, Kirschbaum J, Brückner H (2007) New sequences, constituents, and producers of peptaibiotics: an updated review. Chem Biodivers 4(6):1052–1067. doi:10.1002/cbdv.200790096
Fischbach MA, Clardy J (2007) One pathway, many products. Nat Chem Biol 3(7):353–355. doi:10.1038/nchembio0707-353
Ibrahim A, Yang L, Johnston C, Liu X, Ma B, Magarvey NA (2012) Dereplicating nonribosomal peptides using an informatic search algorithm for natural products (iSNAP) discovery. Proc Natl Acad Sci USA 109(47):19196–19201. doi:10.1073/pnas.1206376109
Iida A, Mihara T, Fujita T, Takaishi Y (1999) Peptidic immunosuppressants from the fungus Trichoderma polysporum. Bioorg Med Chem Lett 9(24):3393–3396. doi:10.1016/S0960-894X(99)00621-6
Koehn FE, Carter GT (2005) The evolving role of natural products in drug discovery. Nat Rev Drug Discov 4(3):206–220. doi:10.1038/nrd1657
Lam KS (2007) New aspects of natural products in drug discovery. Trends Microbiol 15(6):279–289. doi:10.1016/j.tim.2007.04.001
Li JW, Vederas JC (2009) Drug discovery and natural products: end of an era or an endless frontier? Science 325(5937):161–165. doi:10.1126/science.1168243
Mohimani H, Kersten RD, Liu WT, Wang M, Purvine SO, Wu S, Brewer HM, Pasa-Tolic L, Bandeira N, Moore BS, Pevzner PA, Dorrestein PC (2014) Automated genome mining of ribosomal peptide natural products. ACS Chem Biol 9(7):1545–1551. doi:10.1021/cb500199h
Mohimani H, Liu WT, Kersten RD, Moore BS, Dorrestein PC, Pevzner PA (2014) NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J Nat Prod 77(8):1902–1909. doi:10.1021/np500370c
Mohimani H, Liu WT, Mylne JS, Poth AG, Colgrave ML, Tran D, Selsted ME, Dorrestein PC, Pevzner PA (2011) Cycloquest: identification of cyclopeptides via database search of their mass spectra against genome databases. J Proteome Res 10(10):4505–4512. doi:10.1021/pr200323a
Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH (2010) An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov 9(9):719–727. doi:10.1038/nrd3074
Schüller A, Hähnke V, Schneider G (2007) SmiLib v2.0: a Java-Based Tool for Rapid Combinatorial Library Enumeration. QSAR Comb Sci 26(3):407–410. doi:10.1002/qsar.200630101
Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC (2012) Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109(26):E1743–E1752. doi:10.1073/pnas.1203689109
Wright GD (2012) Antibiotics: a new hope. Chem Biol 19(1):3–10. doi:10.1016/j.chembiol.2011.10.019
Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, Wodtke A, de Felicio R, Fenner A, Wong WR, Linington RG, Zhang L, Debonsi HM, Gerwick WH, Dorrestein PC (2013) Molecular networking as a dereplication strategy. J Nat Prod 76(9):1686–1699. doi:10.1021/np400413s
Acknowledgments
This work was funded through a Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery grant (RGPIN 371576-2014) (NAM) and a Joint Programme Initiative on Antimicrobial Resistance funded through the Canadian Institutes of Health Research (NAM). CWJ is funded through a CIHR Doctoral Research Award. NAM is supported by the Canada Research Chairs Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: Natural Product Discovery and Development in the Genomic Era. Dedicated to Professor Satoshi Ōmura for his numerous contributions to the field of natural products.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Johnston, C.W., Connaty, A.D., Skinnider, M.A. et al. Informatic search strategies to discover analogues and variants of natural product archetypes. J Ind Microbiol Biotechnol 43, 293–298 (2016). https://doi.org/10.1007/s10295-015-1675-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1675-9