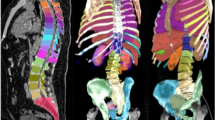
Abstract
Uptake segmentation and classification on PSMA PET/CT are important for automating whole-body tumor burden determinations. We developed and evaluated an automated deep learning (DL)-based framework that segments and classifies uptake on PSMA PET/CT. We identified 193 [18F] DCFPyL PET/CT scans of patients with biochemically recurrent prostate cancer from two institutions, including 137 [18F] DCFPyL PET/CT scans for training and internally testing, and 56 scans from another institution for external testing. Two radiologists segmented and labelled foci as suspicious or non-suspicious for malignancy. A DL-based segmentation was developed with two independent CNNs. An anatomical prior guidance was applied to make the DL framework focus on PSMA-avid lesions. Segmentation performance was evaluated by Dice, IoU, precision, and recall. Classification model was constructed with multi-modal decision fusion framework evaluated by accuracy, AUC, F1 score, precision, and recall. Automatic segmentation of suspicious lesions was improved under prior guidance, with mean Dice, IoU, precision, and recall of 0.700, 0.566, 0.809, and 0.660 on the internal test set and 0.680, 0.548, 0.749, and 0.740 on the external test set. Our multi-modal decision fusion framework outperformed single-modal and multi-modal CNNs with accuracy, AUC, F1 score, precision, and recall of 0.764, 0.863, 0.844, 0.841, and 0.847 in distinguishing suspicious and non-suspicious foci on the internal test set and 0.796, 0.851, 0.865, 0.814, and 0.923 on the external test set. DL-based lesion segmentation on PSMA PET is facilitated through our anatomical prior guidance strategy. Our classification framework differentiates suspicious foci from those not suspicious for cancer with good accuracy.





Similar content being viewed by others
References
Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends—An Update Global Cancer Rates and Trends—An Update. Cancer Epidemiol Biomarkers Prev. 2016; 25:16-27. https://doi.org/10.1158/1055-9965.EPI-15-0578.
Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021; 71:209-249. https://doi.org/10.3322/caac.21660.
Li M, Zelchan R, Orlova A. The Performance of FDA-Approved PET Imaging Agents in the Detection of Prostate Cancer. Biomedicines. 2022;10(10), 2533. https://doi.org/10.3390/biomedicines10102533.
Voter AF, Werner RA, Pienta KJ, et al. Piflufolastat F-18 (18F-DCFPyL) for PSMA PET imaging in prostate cancer. Expert Rev Anticancer Ther. 2022; 22:681-694. https://doi.org/10.1080/14737140.2022.2081155.
Hofman MS, Lawrentschuk N, Francis RJ, et al. Prostate-specific membrane antigen PET-CT in patients with high-risk prostate cancer before curative-intent surgery or radiotherapy (proPSMA): a prospective, randomised, multicentre study. Lancet. 2020; 395:1208-1216. https://doi.org/10.1016/S0140-6736(20)30314-7.
An S, Huang G, Liu J, Wei W. PSMA-targeted theranostics of solid tumors: applications beyond prostate cancers. Eur J Nucl Med Mol Imaging. 2022; 49:3973-3976. https://doi.org/10.1007/s00259-022-05905-7.
Werner RA, Habacha B, Lütje S, et al. High SUVs have more robust repeatability in patients with metastatic prostate cancer: Results from a prospective test-retest cohort imaged with 18F-DCFPyL. Mol Imaging. 2022; 2022:7056983. https://doi.org/10.1155/2022/7056983.
Gafita A, Bieth M, Krönke M, et al. qPSMA: Semiautomatic Software for Whole-Body Tumor Burden Assessment in Prostate Cancer Using 68Ga-PSMA11 PET/CT. J Nucl Med. 2019;60:1277-1283. https://doi.org/10.2967/jnumed.118.224055.
Kim M, Seifert R, Fragemann J, et al. Evaluation of thresholding methods for the quantification of [68Ga] Ga-PSMA-11 PET molecular tumor volume and their effect on survival prediction in patients with advanced prostate cancer undergoing [177Lu]Lu-PSMA-617 radioligand therapy. Eur J Nucl Med Mol Imaging. 2023, 50(7), 2196–2209. https://doi.org/10.1007/s00259-023-06163-x
Karimi D, Dou H, Warfield SK, Gholipour A. Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis. Med Image Anal. 2020; 65:101759. https://doi.org/10.1016/j.media.2020.101759.
Budd S, Robinson EC, Kainz B. A survey on active learning and human-in-the-loop deep learning for medical image analysis. Med Image Anal. 2021; 71:102062. https://doi.org/10.1016/j.media.2021.102062.
Jin W, Li X, Fatehi M, Hamarneh G. Guidelines and evaluation of clinical explainable AI in medical image analysis. Med Image Anal. 2023; 84:102684. https://doi.org/10.1016/j.media.2022.102684.
Kshatri SS, Singh D. Convolutional Neural Network in Medical Image Analysis: A Review. Arch Comput Methods Eng. 2023; 30:2793-2810. https://doi.org/10.1007/s11831-023-09898-w.
Salpea N, Tzouveli P, Kollias D. Medical Image Segmentation: A Review of Modern Architectures. In: Karlinsky, L., Michaeli, T., Nishino, K. (eds) Computer Vision – ECCV 2022 Workshops. ECCV 2022. Lecture Notes in Computer Science, vol 13807. Springer, Cham. https://doi.org/10.1007/978-3-031-25082-8_47.
Kendrick J, Francis R J, Hassan G M, et al. Fully automatic prognostic biomarker extraction from metastatic prostate lesion segmentations in whole-body [68Ga] Ga-PSMA-11 PET/CT images. Eur J Nucl Med Mol Imaging. 2020, 50(1), 67-79. https://doi.org/10.1007/s00259-022-05927-1.
Sibille L, Seifert R, Avramovic N, et al. 18F-FDG PET/CT Uptake Classification in Lymphoma and Lung Cancer by Using Deep Convolutional Neural Networks. Radiology. 2020; 294:445-452. https://doi.org/10.1148/radiol.2019191114.
Yixi X, Ivan K, Sara H, et al. Automatic segmentation of prostate cancer metastases in PSMA PET/CT images using deep neural networks with weighted batch-wise dice loss. Computers in Biology and Medicine. 2023;158:106882. https://doi.org/10.1016/j.compbiomed.2023.106882.
Lindgren Belal S, Frantz S, Minarik D, et al. Applications of Artificial Intelligence in PSMA PET/CT for Prostate Cancer Imaging. Semin Nucl Med. 2023;S0001-2998(23)00049-1. https://doi.org/10.1053/j.semnuclmed.2023.06.001.
Mena E, Rowe SP, Shih JH, et al. Predictors of 18F-DCFPyL PET/CT Positivity in Patients with Biochemical Recurrence of Prostate Cancer After Local Therapy. J Nucl Med. 2022; 63:1184-1190. https://doi.org/10.2967/jnumed.121.262347.
Werner RA, Derlin T, Lapa C, et al. 18F-Labeled, PSMA-Targeted Radiotracers: Leveraging the Advantages of Radiofluorination for Prostate Cancer Molecular Imaging. Theranostics. 2020; 10:1-16. https://doi.org/10.7150/thno.37894.
Rowe SP, Buck A, Bundschuh RA, et al. [18F] DCFPyL PET/CT for imaging of prostate cancer. Nuklearmedizin. 2022; 61:240-246. https://doi.org/10.1055/a-1659-0010.
Yechiel Y, Orr Y, Gurevich K, Gill R, Keidar Z. Advanced PSMA-PET/CT Imaging Parameters in Newly Diagnosed Prostate Cancer Patients for Predicting Metastatic Disease. Cancers. 2023;15(4):1020. https://doi.org/10.3390/cancers15041020.
Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;30. https://doi.org/10.48550/arXiv.1706.03762.
Dosovitskiy A, Beyer L, Kolesnikov A, et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv preprint arXiv:11929 2020. https://doi.org/10.48550/arXiv.2010.11929.
Hatamizadeh A, Tang Y, Nath V, et al. UNETR: Transformers for 3D Medical Image Segmentation. Proceedings of the IEEE/CVF winter conference on applications of computer vision; 2022. p. 574-84. https://doi.org/10.48550/arXiv.2103.10504.
Schmidkonz C, Cordes M, Schmidt D, et al. 68Ga-PSMA-11 PET/CT-derived metabolic parameters for determination of whole-body tumor burden and treatment response in prostate cancer. Eur J Nucl Med Mol Imaging. 2018; 45:1862-1872. https://doi.org/10.1007/s00259-018-4042-z.
Schmuck S, von Klot CA, Henkenberens C, et al. Initial experience with volumetric 68Ga-PSMA I&T PET/CT for assessment of whole-body tumor burden as a quantitative imaging biomarker in patients with prostate cancer. J Nucl Med. 2017; 58:1962-1968. https://doi.org/10.2967/jnumed.117.193581.
Gul S, Khan MS, Bibi A, Khandakar A, Ayari MA, Chowdhury MEH. Deep learning techniques for liver and liver tumor segmentation: A review. Comput Biol Med. 2022; 147:105620. https://doi.org/10.1016/j.compbiomed.2022.105620.
Mohammed YMA, El Garouani S, Jellouli I. A survey of methods for brain tumor segmentation-based MRI images. Finite Elem Anal Des. 2023; 10:266-293. https://doi.org/10.1093/jcde/qwac141.
Park J, Kang SK, Hwang D, et al. Automatic Lung Cancer Segmentation in [18F] FDG PET/CT Using a Two-Stage Deep Learning Approach. Nucl Med Mol Imaging. 2023; 57:86-93. https://doi.org/10.1007/s13139-022-00745-7.
Seifert R, Herrmann K, Kleesiek J, et al. Semiautomatically Quantified Tumor Volume Using 68Ga-PSMA-11 PET as a Biomarker for Survival in Patients with Advanced Prostate Cancer. J Nucl Med. 2020; 61(12):1786-1792. https://doi.org/10.2967/jnumed.120.242057.
Johnsson K, Brynolfsson J, Sahlstedt H, et al. Analytical performance of aPROMISE: automated anatomic contextualization, detection, and quantification of [18F] DCFPyL (PSMA) imaging for standardized reporting. Eur J Nucl Med Mol Imaging. 2022; 49:1041-1051. https://doi.org/10.1007/s00259-021-05497-8.
Nickols N, Anand A, Johnsson K, et al. aPROMISE: A Novel Automated PROMISE Platform to Standardize Evaluation of Tumor Burden in 18F-DCFPyL Images of Veterans with Prostate Cancer. J Nucl Med. 2022; 63:233-239. https://doi.org/10.2967/jnumed.120.261863.
Kendrick J, Francis RJ, Hassan GM. et al. Prognostic utility of RECIP 1.0 with manual and AI-based segmentations in biochemically recurrent prostate cancer from [68Ga] Ga-PSMA-11 PET images. Eur J Nucl Med Mol Imaging. 2023;50:4077-86. https://doi.org/10.1007/s00259-023-06382-2.
36. Capobianco N, Sibille L, Chantadisai M, et al. Whole-body uptake classification and prostate cancer staging in 68Ga-PSMA-11 PET/CT using dual-tracer learning. Eur J Nucl Med Mol Imaging. 2022; 49:517-526. https://doi.org/10.1007/s00259-021-05473-2.
Funding
This study was supported by the Department of Radiology and Radiological Sciences at Johns Hopkins NIH grant U01CA140204, T32EB006351, EB024405, CA134675, and the Science and Technology Innovation Program of Hunan Province (No. 2021RC5003), the National Natural Science Foundation of China (No. 62376287), the International Science and Technology Innovation Joint Base of Machine Vision and Medical Image Processing in Hunan Province number (2021CB1013), the Natural Science Foundation of Hunan Province number (S2024JJQNJJ2617, 2022JJ30762, 2023JJ70016), and the 111 project (B18059).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
M.G.P. is co-inventor on a US patent covering 18F-DCFPyL and as such is entitled to a portion of any licensing fees and royalties generated by this technology. The arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. S.P.R. is a consultant to Progenics Pharmaceuticals, the licensee of 18F-DCFPyL. M.G.P and S.P.R receive research funding from Progenics Pharmaceuticals. No other potential conflicts of interest relevant to this article exist.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Imami, M.R., Zhao, L. et al. An Automated Deep Learning-Based Framework for Uptake Segmentation and Classification on PSMA PET/CT Imaging of Patients with Prostate Cancer. J Digit Imaging. Inform. med. (2024). https://doi.org/10.1007/s10278-024-01104-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10278-024-01104-y