Abstract
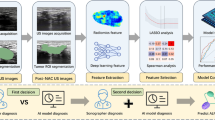
The paper aims to develop prediction model that integrates clinical, radiomics, and deep features using transfer learning to stratifying between high and low risk of thymoma. Our study enrolled 150 patients with thymoma (76 low-risk and 74 high-risk) who underwent surgical resection and pathologically confirmed in Shengjing Hospital of China Medical University from January 2018 to December 2020. The training cohort consisted of 120 patients (80%) and the test cohort consisted of 30 patients (20%). The 2590 radiomics and 192 deep features from non-enhanced, arterial, and venous phase CT images were extracted and ANOVA, Pearson correlation coefficient, PCA, and LASSO were used to select the most significant features. A fusion model that integrated clinical, radiomics, and deep features was developed with SVM classifiers to predict the risk level of thymoma, and accuracy, sensitivity, specificity, ROC curves, and AUC were applied to evaluate the classification model. In both the training and test cohorts, the fusion model demonstrated better performance in stratifying high and low risk of thymoma. It had AUCs of 0.99 and 0.95, and an accuracy of 0.93 and 0.83, respectively. This was compared to the clinical model (AUCs of 0.70 and 0.51, accuracy of 0.68 and 0.47), the radiomics model (AUCs of 0.97 and 0.82, accuracy of 0.93 and 0.80), and the deep model (AUCs of 0.94 and 0.85, accuracy of 0.88 and 0.80). The fusion model integrating clinical, radiomics and deep features based on transfer learning was efficient for noninvasively stratifying high risk and low risk of thymoma. The models could help to determine surgery strategy for thymoma cancer.






Similar content being viewed by others
Data Availability
The dataset from Shengjing hospital was used under approval for the current study. Restrictions apply to the availability of this dataset and so it is not publicly available.
Abbreviations
- LASSO:
-
Least absolute shrinkage and selection operator
- SVM:
-
Support vector machines
- CI:
-
Confidence intervals
- VGG:
-
Visual geometry group
- ROC:
-
Receiver operating characteristic curve
- AUC:
-
Area under curve
- DCA:
-
Decision curve analysis
- ANOVA:
-
Analysis of variance
References
https://www.cancer.org/cancer/thymus-.cancer/detection-diagnosis-staging/survival-rates.html.
Travis WD, Burke, et al. WHO Classification of Tumors of the Lung, Pleura, Thymus and Heart. 4 edition[J].
Jeong YJ, Lee KS, Kim J, et al. Does CT of Thymic Epithelial Tumors Enable Us to Differentiate Histologic Subtypes and Predict Prognosis?[J]. American Journal of Roentgenology, 2004, 183(2): 283- 289.
Motohiko Y, Kanako O, Hajime U, et al. Quantitative 3D Shape Analysis of CT Images of Thymoma: A Comparison With Histological Types[J].AJR AM J Roentgenol, 2020, 214(2):341–347.
Lee H S, Oh J S, Park Y S, et al. Differentiating the grades of thymic epithelial tumor malignancy using textural features of intratumoral heterogeneity via18F-FDG PET/CT[J]. Annals of Nuclear Medicine, 2016, 30(4):309-319.
Yasaka K, Akai H, Nojima M, et al. Quantitative computed tomography texture analysis for estimating histological subtypes of thymic epithelial tumors[J].European Journal of Radiology, 2017:S0720048X17301651.
Li B, Xin Y K, Xiao G, et al. Predicting pathological subtypes and stages of thymic epithelial tumors using DWI: value of combining ADC and texture parameters[J].European Radiology, 2019, 29(10):5330–5340
Xiao G, Rong W C, Hu Y C, et al. MRI Radiomics Analysis for Predicting the Pathologic Classification and TNM Staging of Thymic Epithelial Tumors: A Pilot Study[J]. AJR AM J Roentgenol, 2020, 214(2):328-340.
Ayten Kayi Cangir et al, CT imaging-based machine learning model: a potential modality for predicting low-risk and high-risk groups of thymoma: “Impact of surgical modality choice”. World Journal of Surgical Oncology (2021) 19:147.
Wentao Dong et al, Application of a combined radiomics nomogram based on CE-CT in the preoperative prediction of thymomas risk categorization. Frontiers in Oncology, (2022), https://doi.org/10.3389/fonc.2022.944005).
Jin Liu et al, CT-Based Radiomics Signatures for Predicting the Risk Categorization of Thymic Epithelial Tumors. Frontiers in Oncology, (2021), https://doi.org/10.3389/fonc.2021.628534.
Lan Shang et al, Machine-learning classifiers based on non-enhanced computed tomography radiomics to differentiate anterior mediastinal cysts from thymomas and low-risk from high-risk thymomas: A multicenter study. Frontiers in Oncology, 2022. https://doi.org/10.3389/fonc.2022.1043163
Chunhai Yu et al, Contrast-enhanced CT-based radiomics model for diferentiating risk subgroups of thymic epithelial tumors. BMC Medical Imaging, 2022, 22:37. https://doi.org/10.1186/s12880-022-00768-8.
Zhong, Y. et al. Radiomics approach to prediction of occult mediastinal lymph node metastasis of lung adenocarcinoma. AJR Am. J. Roentgenol. 2018,211:109–113
Walsh TN, Noonan N, Hollywood D, Kelly A, Keeling N, Hennessy TP. A comparison of multimodal therapy and surgery for esophageal adenocarcinoma. N Engl J Med. 1996;335(7):462-467.
Dey N, et al. Customized VGG19 Architecture for Pneumonia Detection in Chest X-Rays. PATTERN RECOGNITION LETTERS, (2021):67–74.
Skrede OJ, De Raedt S, Kleppe A, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 2020; 395: 350–60.
Shin, H. C. et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 35, 1285–1298 (2016).
Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016; 34: 2157–64.
Chang, C. C. & Lin, C. J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 1–27 (2011).
Ueno H, Ishiguro M, Nakatani E, et al. Prospective multicenter study on the prognostic and predictive impact of tumor budding in stage II colon cancer: results from the SACURA trial. J Clin Oncol 2019; 37: 1886–94.
Wenyu Gao et al, A predictive model integrating deep and radiomics features based on gadobenate dimeglumine‑enhanced MRI for postoperative early recurrence of hepatocellular carcinoma. La radiologia medica (2022) 127:259–271, https://doi.org/10.1007/s11547-021-01445-6.
Xueyi Zheng et al, Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. NATURE COMMUNICATIONS, https://doi.org/10.1038/s41467-020-15027-z
Wei Wang, MD, PhD, et al. Development and Validation of a Computed Tomography–Based Radiomics Signature to Predict Response to Neoadjuvant Chemotherapy for Locally Advanced Gastric Cancer. JAMA network open 2021 4(8).
D. Dong et al, Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multicenter study. Annals of oncology 2021 31:912-920.
Song D et al, Using deep learning to predict microvascular invasion in hepatocellular carcinoma based on dynamic contrast-enhanced MRI combined with clinical parameters. J Cancer Res Clin Oncol. https://doi.org/10.1007/s00432.021.03617.3
Ning Z et al, Pattern classification for gastrointestinal stromal tumors by integration of radiomics and deep convolutional features. IEEE J Biomed Health Inform 23(3):1181–1191. https://doi.org/10.1109/JBHI.2018.2841992
Paul R et al, Predicting malignant nodules by fusing deep features with classical radiomics features. J Med Imaging (Bellingham) 5(1):011021. https://doi.org/10.1117/1.JMI.5.1.011021
Xueyi Zheng et al, Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nature communications 2020. https://doi.org/10.1007/s00330-021-08237-6.
Ruitian Gao et al, Deep learning for differential diagnosis of malignant hepatic tumors based on multi-phase contrast-enhanced CT and clinical data. J Hematol Oncol, (2021) 144:154. https://doi.org/10.1186/s13045-021-01167-2.
Funding
The study was supported by natural science funding project of education department of Liaoning Province, China (No. LJKMZ20221196).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Points
• The study introduces a novel method for clinical diagnosis of thymoma risk based on transfer learning technology.
• An integrated analysis of multi-omics and multi-modal features provided a strong foundation for a deeper understanding of tumor risk.
• The fusion model was efficient for noninvasively stratifying high and low risk of thymoma with an accuracy of 0.83, an AUC of 0.95, a sensitivity of 0.80 and a specificity of 1.00.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, W., Wang, W., Zhang, H. et al. Development and Validation of Multi-Omics Thymoma Risk Classification Model Based on Transfer Learning. J Digit Imaging 36, 2015–2024 (2023). https://doi.org/10.1007/s10278-023-00855-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-023-00855-4