Abstract
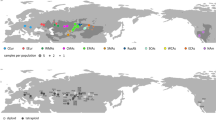
A group of temperate grassland plant species termed the “Mansen elements” occurs in Japan and is widely distributed in the grasslands of continental East Asia. It has been hypothesized that these species are continental grassland relicts in Japan that stretch back to a colder age, but their migration history has not been elucidated. To assess the migration history of the Mansen elements, we performed phylogeographic analyses of Tephroseris kirilowii, a member of this group, using single-nucleotide polymorphisms (SNPs) obtained from multiplexed inter-simple sequence repeat genotyping by sequencing (MIG-seq). It was estimated that the Japanese populations of T. kirilowii were divided from those of continental East Asia at 25.2 thousand years ago (ka) with 95% highest probability density interval (HPD) of 15.3–40.0 ka and that Japanese clades first diverged at 20.2 ka with 95% HPD of 10.4–30.1 ka. As the climatically suitable range during the last glacial maximum (LGM) estimated using ecological niche modeling (ENM) was limited in Japan and there was a slight genetic differentiation among Japanese populations, a post-glacial expansion of T. kirilowii in the Japanese Archipelago was indicated.








Similar content being viewed by others
Data Availability
It is described in the M & M part as "These nucleotide sequence data were deposited in the DDBJ database with the accession number of DRA015152."
References
Arizono S (1995) Land use in Japan circa 1850. In: Himiyama Y, Arai T, Ota I, Kubo S, Tamura T, Nogami M, Murayama Y, Yorifuji T (eds) Atlas. Environmental change in modern Japan. Asakura-shoten, Tokyo, pp. 4–5 (in Japanese)
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH (2011) Stacks : building and genotyping loci de novo from short-read sequences. G3 1: 171–182. https://doi.org/10.1534/g3.111.000240
Cornuet J-M, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Mariin J-M, Estoup A (2014) DIYABC v2.0: a software to make approximate bayesian computation inferences about population history using single nucleotide polymorphism. DNA Sequence Microsatellite Data Bioinformat 30:1187–1189. https://doi.org/10.1093/bioinformatics/btt763
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, vonHoldt BM (2012) Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Gent PP, Danabasoglu G, Donner LJ, Holland MM, Hunke EC, Jayne SR, Lawrence DM, Neale RB, Rasch PJ, Vertenstein M, Worley PH, Yang Z-L, Zhang M (2011) The community climate system model version 4. J Climate 24:4973–4991. https://doi.org/10.1175/2011JCLI4083.1
Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167:747–760. https://doi.org/10.1534/genetics.103.024182
Hey J, Nielsen R (2007) Integration within the felsenstein equation for improved markov chain monte carlo methods in population genetics. Proc Natl Acad Sci USA 104:2785–2790. https://doi.org/10.1073/pnas.0611164104
Hey J, Chung Y, Sethuraman A, Lachance J, Tishkoff S, Sousa VC, Wang Y (2018) Phylogeny estimation by integration over isolation with migration models. Mol Biol Evol 35:2805–2818. https://doi.org/10.1093/molbev/msy162
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978. https://doi.org/10.1002/joc.1276
Hotta M (1974) History and geography of plants. Sanseido, Tokyo (in Japanese)
Kadota Y (2017) Tephroseris. In: Ohashi H, Kadota Y, Murata J, Yonekura K, Kihara H (eds) Wild flowers of Japan 5. Heibonsha, Tokyo, pp 311–312 (in Japanese)
Kamada M, Nakagoshi N (1997) Influence of cultural factors on landscapes of mountainous farm villages in Japan. Landscape Urban Planning 37:85–90
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. https://doi.org/10.1093/bib/bbx108
Kitamura S (1957) Distribution of Plants. In: Kitamura S, Murata G, Hori M (eds) Coloured illustrations of herbaceous plants of Japan I. Hoikusha, Osaka, pp 246–264 (in Japanese)
Kitamura S (1981) Senecio. In: Satake Y, Ohwi J, Kitamura S, Watari S, Tominari T (eds) Wild flowers of Japan, herbaceous plants 3. Heibonsha, Tokyo, pp 181–183 (in Japanese)
Kiyokawa S, Ito T, Ikehara M, Onoue T (2014) Super time scale of the earth. Iwanami-shoten, Tokyo
Koidzumi G (1931) Zengen. In: Maebara K (ed) Florula austro-higoensis. Sanshusha, Tokyo, pp. xv–xviii (in Japanese)
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak : a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Koyama H (1995) Senecio. In: Yamazaki T, Boufford DE, Ohba H (eds) aIwatsuki K. Flora of Japan IIIb. Kodansha, Tokyo, pp 40–44
Leigh JW, Bryant D (2015) popart : full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210X.12410
Luu K, Bazin E, Blum MGB (2017) pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol Ecol Resour 17:67–77. https://doi.org/10.1101/056135
Miyabychi Y, Sugiyama S, Nagaoka Y (2012) Vegetation and fire history during the last 30,000 years based on phytolith and macroscopic charcoal records in the eastern and western areas of Aso Volcano, Japan. Quatern Int 254:28–35
Murata G (1977) Phytogeographical consideration on the flora and vegetation of Japan. Acta Phytotax Geobot 28:65–83 (in Japanese)
Murata G (1988) The distribution on the continental elements in Japan. Flora and its characteristic in Japan 17. Nihon No Seibutsu 2:21–25 (in Japanese)
Nakahama N, Uchida K, Ushimaru A, Isagi Y (2018) Historical changes in grassland area determined the demography of semi-natural grassland butterflies in Japan. Heredity 121:155–168
Noshiro S (2005) Vegetation history in Japan. In: Fukushima T, Iwase T (eds) Illustrated vegetation of Japan. Asakura-shoten, Tokyo, pp 12–15 (in Japanese)
Numata M (1961) Ecology of grasslands in Japan. J College Arts Sciences, Chiba Univ 3:327–345
Oh SY, Pak JH (2001) Distribution maps of vascular plants in Korea. Academybook Publishing, Seoul (in Korean)
Okamura M, Matsufuji K, Kimura H, Tsuji S, Baba H (1998) Paleolithic archeology. Gakuseisha, Tokyo (in Japanese)
Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94. https://doi.org/10.1126/science.1180677
Otto-Bliesner BL, Marshall SJ, Overpeck JT, Miller GH, Hu A, CAPE Last Interglacial Project members (2006) Simulating arctic climate warmth and icefield retreat in the Last Interglaciation. Science 311:1751–1753. https://doi.org/10.1126/science.1120808
Pfeifer B, Wittelsburger U, Ramos-Onsins SE, Lercher MJ (2014) PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol 31:1929–1936. https://doi.org/10.1093/molbev/msu136
Phillips SJ, Anderson RP, Dudík M, Schapire RE, Blair ME (2017) Opening the black box: an open-source release of Maxent. Ecography 40:887–893. https://doi.org/10.1111/ecog.03049
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data 15.
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Sakaguchi Y (1987) Japanese prehistoric culture flourished in forest-grassland mixd area. Bull Dept Geogr Univ Tokyo 19:1–19
Sata H, Shimizu M, Iwasaki T, Ikeda H, Soejima A, Kozhevnikov AE, Kozhevnikova ZV, Im H-T, Jang S-K, Azuma T, Nagano AJ, Fujii N (2021) Phylogeography of the East Asian grassland plant, Viola orientalis (Violaceae), inferred from plastid and nuclear restriction site-associated DNA sequencing data. J Plant Res 134:1181–1198. https://doi.org/10.1007/s10265-01339-8
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Suka T (2012) Semi-natural Grasslands in the Japanese Archipelago. In: Suka T, Okamoto T, Ushimaru A (eds) Grassland and Japanese. Tsukiji-shokan, Tokyo (in Japanese)
Suka T, Ushimaru A, Tanaka H (2011) Grassland history and grassland fauna and flora in the Japanese Archipelago. In: Yumoto T (ed) Environmental history of grassfields in Japan. Bun-ichi, Tokyo, pp 101–122 (in Japanese)
Suyama Y, Matsuki Y (2015) MIG-seq: an effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci Rep 5:16963. https://doi.org/10.1038/srep16963
Suyama Y, Hirota SK, Matsuo A, Tsunamoto Y, Mitsuyuki C, Shimura A, Okano K (2022) Complementary combination of multiplex high-throughput DNA sequencing for molecular phylogeny. Ecol Res 37:171–181
Tabata H (1997) Meadow plants in Satoyama. In: Tabata H (ed) Satoyama and its conservation. Hoikusha, Osaka, pp 38–41 (in Japanese)
Takaishi A, Kozhevnikov AE, Kozhevnikova ZV, Ikeda H, Fujii N, Soejima A (2019) Phylogeography of Pulsatilla cernua (Ranunculaceae), a grassland species, in Japan. Ecol Evol 9:7262–7272. https://doi.org/10.1002/ece3.5298
Tanabe AS, Toju H (2013) Two new computational methods for universal DNA barcoding: A benchmark using barcode sequences of bacteria, archaea animals, fungi, and land plants. PLoS ONE. https://doi.org/10.1371/journal.pone.0076910
Ushimaru A, Uchida K, Suka T (2018) Grassland biodiversity in Japan: threats, management and conservation. In: Squires VR, Dengler J, Feng H, Hua L (eds) Grasslands of the world. CRC Press Taylor and Francis Group, Boca Raton, London, New York, pp 197–218
Watanabe S, Hajima T, Sudo K, Nagashima T, Takemura T, Okajima H, Nozawa T, Kawase H, Abe M, Yokohata T, Ise T, Sato H, Kato E, Takata K, Emori S, Kawamiya M (2011) MIROC-ESM 2010: model description and basic results of CMIP5-20c3m experiments. Geosci Model Dev 4:845–872. https://doi.org/10.5194/gmd-4-845-2011
Yamaura Y, Oka H, Taki H, Ozaki K, Tanaka H (2012) Sustainable management of planted landscapes: lessons from Japan. Biodivers Conserv 21:3107–3129. https://doi.org/10.1007/s10531-012-0357-4
Yamaura Y, Narita A, Kusumoto Y, Nagano AJ, Tezuka A, Okamoto T, Takahara H, Nakamura F, Isagi Y, Lindenmayer D (2019) Genomic reconstruction of 100 000-year grassland history in a forested country: population dynamics of specialist forbs. Biol Lett 15:20180577. https://doi.org/10.1098/rsbl.2018.0577
Yiling C, Nordenstam B, Jeffrey C, (2011) Tephroseris. In: Wu ZY, Raven PH, Hong DY (eds) Flora of China vol 20–21 (Asteraceae). Science Press, Beijing and Missouri Botanical Garden Press (St. Louis). pp. 481–487.
Acknowledgements
The authors are grateful to Koya Akiyama, Chengyong Guo, Sachiko Horie, Shungo Kariyama, Takahide Kurosawa, Masayuki Maki, Kiyotaka Minoda, Yuki Mitsui, Shuichi Nemoto, Yudai Okuyama, Wataru Onishi, Yoko Ota, Sumio Sei, Kotaro Shuto, Shuichiro Tagane, Kikou Takamura, Michihisa Takashima, Kiyoto Takazoe, Yuki Tanabe, Atsushi Ushimaru, and Shigeaki Yatagai (alphabetical order, title omitted) for their assistance in sample collection. We thank the herbaria, KANA, SAPS, and Kumamoto Prefecture Museum Network Center for providing materials. This study was supported by JSPS KAKENHI Grant Number JP17H03721 and a grant from the Institute of Plant Science and Resources, Okayama University (3037, 3140, and R236). All the authors have no conflict of interest, financial or otherwise.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sakaba, T., Soejima, A., Fujii, S. et al. Phylogeography of the temperate grassland plant Tephroseris kirilowii (Asteraceae) inferred from multiplexed inter-simple sequence repeat genotyping by sequencing (MIG-seq) data. J Plant Res 136, 437–452 (2023). https://doi.org/10.1007/s10265-023-01452-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-023-01452-w