Abstract
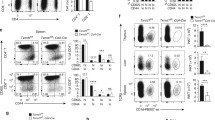
CYtochrome P450, family 51 (CYP51) is an important enzyme for de novo cholesterol synthesis in mammalian cells. In the present study, we found that the expression of CYP51 positively correlated with CD4+ T cell activation both in vivo and in vitro. The addition of ketoconazole, a pharmacological inhibitor of CYP51, prevented the proliferation and activation of anti-CD3/CD28-expanded mouse CD4+ T cells in a dose-dependent fashion. Liquid chromatography-tandem mass spectrometry indicated an increase in levels of lanosterol in T cells treated with ketoconazole during activation. Ketoconazole-induced blockade of the cholesterol synthesis pathway also caused Sterol regulatory element binding protein 2 (SREBP2) activation in CD4+ T cells. Additionally, ketoconazole treatment elicited an integrated stress response in T cells that up-regulated activating transcription factor 4 (ATF4) and DNA-damage inducible transcript 3 (DDIT3/CHOP) at the translational level. Furthermore, treatment with ketoconazole significantly decreased the amount of CD4+ T cells infiltrating lesions in the submandibular glands of NOD/Ltj mice. In summary, our results suggest that CYP51 plays an essential role in the proliferation and survival of CD4+ T cells, which makes ketoconazole an inhibitor of CD4+ T cell proliferation and of the SS-like autoimmune response through regulating the biosynthesis of cholesterol and inducing the integrated stress response.







Similar content being viewed by others
Availability of data and materials
Data banks/repositories corresponding to all datasets analyzed in this study were listed in Additional file 1.
Abbreviations
- ABCA1:
-
ATP-binding cassette transporter A 1
- ABCG1:
-
ATP-binding cassette transporter G 1
- ATF4:
-
Activating transcription factor 4
- BIM:
-
Bcl-2 interacting mediator of cell death
- BP:
-
Biological process
- CDK2:
-
Cyclin-dependent kinase 2
- CDK4:
-
Cyclin-dependent kinase 4
- CFSE:
-
5-(And -6) -Carboxyfluorescein Daicetate Succinimidyl Ester
- CYP51:
-
Lanosterol 14alpha-demethylase
- Cyp51:
-
Cytochrome P450 family 51
- CYP51A1:
-
Cytochrome P450 family 51 subfamily A member 1
- Ddit3:
-
DNA-damage inducible transcript 3
- DEGs:
-
Differentially expressed genes
- DMSO:
-
Dimethyl sulfoxide
- Eif2ak3:
-
Eukaryotic translation initiation factor 2 alpha kinase 3
- FDPS:
-
Farnesyl pyrophosphate synthase
- FBS:
-
Fetal bovine serum
- GEO:
-
Gene Expression Omnibus database
- GO:
-
Gene Ontology
- GSEA:
-
Gene Set Enrichment Analysis
- H&E:
-
Hematoxylin & eosin
- HMGCR:
-
Hydroxymethylglutaryl-CoA reductase
- HMG-CoA:
-
3-Hydroxy-3-methyl glutaryl coenzyme A reductase
- IFN-γ:
-
Interferon-γ
- IL-17:
-
Interleukin-17
- IL-2:
-
Interleukin-2
- HMGCR:
-
Hydroxymethylglutaryl-CoA reductase
- IDI1:
-
Isopentenyl pyrophosphate isomerase 1
- IL-6:
-
Interleukin-6
- JNK/STAT:
-
Janus kinase/signal transducer and activator of transcription
- KTC:
-
Ketoconazole
- LC–MS:
-
Liquid chromatography-tandem mass spectrometry
- ISR:
-
Integrated stress response
- LXRβ:
-
Liver x receptor β
- MBCD:
-
Methyl-β-cyclodextrin
- MCL-1:
-
Myeloid cell leukemia-1
- mRNA:
-
Messenger RNAs
- NCBI:
-
National Center for Biotechnology Information
- p38MAPK:
-
P38 mitogen-activated protein kinase
- PBS:
-
Phosphate buffered saline
- PCR:
-
Polymerase chain reaction
- PBMC:
-
Peripheral blood mononuclear cell
- PERK:
-
Protein kinase RNA–like endoplasmic reticulum kinase
- RA:
-
Rheumatoid arthritis
- SEM:
-
Standard deviations of the mean
- SFR:
-
Salivary flow rate
- siRNA:
-
Small-hairpin RNA
- SLE:
-
Systematic lupus erythematosus
- SQLE:
-
Squalene epoxidase
- SREBP2:
-
Sterol regulatory element binding protein2
- SS:
-
Sjogren’s syndrome
- Th1:
-
T helper 1
- TNF-α:
-
Tumor necrosis factor-α
- 7-DHC:
-
7-Dehydrocholesterol
References
Mavragani CP, Moutsopoulos HM. Sjögren’s syndrome. Annu Rev Pathol Mech Dis. 2014;9:273–85.
Fox RI. Sjögren’s syndrome. In: Lancet. Elsevier, 2005 p. 321–331
Tang Y, Zhou T, Yu X, Xue Z, Shen N. The role of long non-coding RNAs in rheumatic diseases. Nat Rev Rheumatol. 2017;13:657–69.
Malladi AS, Sack KE, Shiboski SC, et al. Primary Sjögren’s syndrome as a systemic disease: a study of participants enrolled in an international Sjögren’s syndrome registry. Arthritis Care Res. 2012;64:911–8.
Singh N, Cohen PL. The T cell in Sjogren’s syndrome: force majeure, not spectateur. J Autoimmun. 2012;39:229–33.
Wei L, Zhifei X, Xiaoran N, et al. Patients with early-onset primary Sjögren’s syndrome have distinctive clinical manifestations and circulating lymphocyte profiles. Rheumatology (Oxford). 2022;61:597–605.
Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–30.
Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445–89.
da Silva TA, Oliveira-Brito PKM, Gonçalves TE, Vendruscolo PE, Roque-Barreira MC. ArtinM mediates murine T cell activation and induces cell death in Jurkat human leukemic T cells. Int J Mol Sci. 2017;18(7):1400.
Hohlfeld R, Dornmair K, Meinl E, Wekerle H. The search for the target antigens of multiple sclerosis, part 1: autoreactive CD4+ T lymphocytes as pathogenic effectors and therapeutic targets. Lancet Neurol. 2016;15:198–209.
Nocturne G, Mariette X. Advances in understanding the pathogenesis of primary Sjögren’s syndrome. Nat Rev Rheumatol. 2013;9:544–56.
Van Woerkom JM, Kruize AA, Wenting-Van Wijk MJG, et al. Salivary gland and peripheral blood T helper 1 and 2 cell activity in Sjögren’s syndrome compared with non-Sjögren’s sicca syndrome. Ann Rheum Dis. 2005;64:1474–9.
Roescher N, Tak PP, Illei GG. Cytokines in Sjögren’s syndrome: potential therapeutic targets. Ann Rheum Dis. 2010;69:945–8.
Iwakura Y, Ishigame H, Saijo S, Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–62.
Pérez P, Kwon YJ, Alliende C, et al. Increased acinar damage of salivary glands of patients with Sjögren’s syndrome is paralleled by simultaneous imbalance of matrix metalloproteinase 3/tissue inhibitor of metalloproteinases 1 and matrix metalloproteinase 9/tissue inhibitor of metalloprotein. Arthritis Rheum. 2005;52:2751–60.
Fogli LK, Sundrud MS, Goel S, et al. T cell-derived IL-17 mediates epithelial changes in the airway and drives pulmonary neutrophilia. J Immunol. 2013;191:3100–11.
Shi H, Cao N, Pu Y, Xie L, Zheng L, Yu C. Long non-coding RNA expression profile in minor salivary gland of primary Sjögren’s syndrome. Arthritis Res Ther. 2016;18:109.
Fu J, Shi H, Wang B, et al. LncRNA PVT1 links Myc to glycolytic metabolism upon CD4+ T cell activation and Sjögren’s syndrome-like autoimmune response. J Autoimmun. 2020;107:102358.
Fu J, Pu Y, Wang B, et al. Pharmacological inhibition of glutaminase 1 normalized the metabolic state and CD4+ T cell response in Sjogren’s syndrome. J Immunol Res. 2022;1–13:2022.
Castrejón-Morales CY, Granados-Portillo O, Cruz-Bautista I, et al. Omega-3 and omega-6 fatty acids in primary Sjögren’s syndrome: clinical meaning and association with inflammation. Clin Exp Rheumatol. 2020;38(Suppl 1):34–9.
Lu J, Guo Y, Lu Y, et al. Untargeted lipidomics reveals specific lipid abnormalities in Sjögren’s syndrome. Rheumatology (Oxford). 2021;60:1252–9.
Edgar R. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
Zeng D, Ye Z, Shen R, et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front Immunol. 2021;12:1–9.
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015;1:417–25.
Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16:284–7.
Bruserud Ø. Effects of azoles on human acute myelogenous leukemia blasts and T lymphocytes derived from acute leukemia patients with chemotherapy-induced cytopenia. Int Immunopharmacol. 2001;1:2183–95.
Guide for the care and use of laboratory animals. National academies press, Washington, D.C., Washington (DC), 2011.
Chisholm DM, Waterhouse JP, Mason DK. Lymphocytic sialadenitis in the major and minor glands: a correlation in postmortem subjects. J Clin Pathol. 1970;23:690–4.
Gu F, Xu S, Zhang P, et al. CP-25 Alleviates experimental Sjögren’s syndrome features in NOD/Ltj mice and modulates T lymphocyte subsets. Basic Clin Pharmacol Toxicol. 2018;123:423–34.
Yin J, Zheng Z, Zeng X, et al. lncRNA MALAT1 mediates osteogenic differentiation of bone mesenchymal stem cells by sponging miR-129-5p. PeerJ. 2022;10:e13355.
King RJ, Singh PK, Mehla K. The cholesterol pathway: impact on immunity and cancer. Trends Immunol. 2022;43:78–92.
Bietz A, Zhu H, Xue M, Xu C. Cholesterol metabolism in T cells. Front Immunol. 2017;8:16–26.
Yin J, Zeng X, Ai Z, Yu M, Wu Y, Li S. Construction and analysis of a lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in oral cancer. BMC Med Genomics. 2020;13:1–14.
Zhao Y, Chen D, Yin J, Xie J, Sun C-Y, Lu M. Comprehensive analysis of tumor immune microenvironment characteristics for the prognostic prediction and immunotherapy of oral squamous cell carcinoma. Front Genet. 2022;13:788580.
Hubler Z, Allimuthu D, Bederman I, et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature. 2018;560:372–6.
Li M-X, Yang Y, Zhao Q, et al. Degradation versus inhibition: development of proteolysis-targeting chimeras for overcoming statin-induced compensatory upregulation of 3-hydroxy-3-methylglutaryl coenzyme a reductase. J Med Chem. 2020;63:4908–28.
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66.
Lefever S, Anckaert J, Volders P-J, Luypaert M, Vandesompele J, Mestdagh P. DecodeRNA- predicting non-coding RNA functions using guilt-by-association. Database (Oxford) 2017, 2017.
Khadirnaikar S, Narayanan SP, Shukla SK. Decoding the LncRNA transcriptome of esophageal cancer: identification of clinically relevant LncRNAs. Biomark Med. 2018;12:1083–93.
Guo S, Jian L, Tao K, Chen C, Yu H, Liu S. Novel breast-specific long non-coding RNA LINC00993 acts as a tumor suppressor in triple-negative breast cancer. Front Oncol. 2019;9:1325.
Matsuura S, Fujii-Kuriyama Y, Tashiro Y. Immunoelectron microscope localization of cytochrome P-450 on microsomes and other membrane structures of rat hepatocytes. J Cell Biol. 1978;78:503–19.
Bai LL, Chen H, Zhou P, Yu J. Identification of tumor necrosis factor-alpha (TNF-α) inhibitor in rheumatoid arthritis using network pharmacology and molecular docking. Front Pharmacol. 2021;12:1–12.
Gnedenko OV, Kaluzhskiy LA, Mol’nar AA, et al. SPR-biosensor assay for analysis of small compounds interaction with human cytochrome P450 51A1 (CYP51A1). Biomed Khim. 2013;59(4):388–98. https://doi.org/10.18097/pbmc20135904388.
Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005;1:179–89.
Hong WC, Amara SG. Membrane cholesterol modulates the outward facing conformation of the dopamine transporter and alters cocaine binding. J Biol Chem. 2010;285:32616–26.
Idowu JY, Hagenbuch B. Free cholesterol affects the function and localization of human Na(+)/taurocholate cotransporting polypeptide (NTCP) and organic cation transporter 1 (OCT1). Int J Mol Sci. 2022;23(15):8457.
Wilfahrt D, Philips RL, Lama J, et al. Histone deacetylase 3 represses cholesterol efflux during CD4(+) T-cell activation. Elife. 2021;10:e70978.
Marty-Roix R, Vladimer GI, Pouliot K, et al. Identification of QS-21 as an inflammasome-activating molecular component of saponin adjuvants. J Biol Chem. 2016;291:1123–36.
Kidani Y, Elsaesser H, Hock MB, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–99.
Cui G, Qin X, Wu L, et al. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J Clin Invest. 2011;121:658–70.
Webb LM, Sengupta S, Edell C, et al. Protein arginine methyltransferase 5 promotes cholesterol biosynthesis-mediated Th17 responses and autoimmunity. J Clin Invest. 2020;130:1683–98.
Bensinger SJ, Bradley MN, Joseph SB, et al. LXR Signaling Couples Sterol Metabolism to Proliferation in the Acquired Immune Response. Cell. 2008;134:97–111.
Leichner GS, Avner R, Harats D, Roitelman J. Metabolically regulated endoplasmic reticulum-associated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase: evidence for requirement of a geranylgeranylated protein. J Biol Chem. 2011;286:32150–61.
Harding HP, Zhang Y, Khersonsky S, et al. Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab. 2005;2:361–71.
Zheng M, Zhang Q, Joe Y, et al. Curcumin induces apoptotic cell death of activated human CD4 + T cells via increasing endoplasmic reticulum stress and mitochondrial dysfunction. Int Immunopharmacol. 2013;15:517–23.
Chang SH, Chung Y, Dong C. Vitamin D suppresses Th17 cytokine production by inducing C/EBP Homologous Protein (CHOP) expression. J Biol Chem. 2010;285:38751–5.
Sarmanova A, Doherty M, Kuo C, et al. Erratum: statin use and risk of joint replacement due to osteoarthritis and rheumatoid arthritis: a propensity-score matched longitudinal cohort study. Rheumatology. 2020;59:3120. https://doi.org/10.1093/rheumatology/keaa223.
Li GM, Zhao J, Li B, Zhang XF, Ma JX, Ma XL, Liu J. The anti-inflammatory effects of statins on patients with rheumatoid arthritis: a systemic review and meta-analysis of 15 randomized controlled trials. Autoimmun Rev. 2018;17:215–25.
Chataway J, Schuerer N, Alsanousi A, et al. Eff ect of high-dose simvastatin on brain atrophy and disability in secondary progressive multiple sclerosis (MS-STAT): a randomised, placebo-controlled, phase 2 trial. Lancet. 2014;383:2213–21.
Bogie JFJ, Vanmierlo T, Vanmol J, et al. Liver X receptor beta deficiency attenuates autoimmune-associated neuroinflammation in a T cell-dependent manner. J Autoimmun. 2021;124:102723.
Berghoff SA, Gerndt N, Winchenbach J, et al. Dietary cholesterol promotes repair of demyelinated lesions in the adult brain. Nat Commun. 2017;8(1):1–15.
Fessler MB. The intracellular cholesterol landscape: dynamic integrator of the immune response. Trends Immunol. 2016;37:819–30.
Pawelec G, Ehninger G, Rehbein A, Schaudt K, Jaschonek K. Comparison of the immunosuppressive activities of the antimycotic agents itraconazole, fluconazole, ketoconazole and miconazole on human T-cells. Int J Immunopharmacol. 1991;13:299–304.
Friccius H, Pohla H, Adibzadeh M, Siegels-Hübenthal P, Schenk A, Pawelec G. The effects of the antifungal azoles itraconazole, fluconazole, ketoconazole and miconazole on cytokine gene expression in human lymphoid cells. Int J Immunopharmacol. 1992;14:791–9.
Zhu J, Yamane H, Paul WE. Differentiation of effector CD4+ T cell populations. Annu Rev Immunol. 2010;28:445–89.
Bietz A, Zhu H, Xue M, Xu C. Cholesterol metabolism in T cells. Front Immunol. 2017;8:1–8.
Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. MTORC1 couples immune signals and metabolic programming to establish T reg-cell function. Nature. 2013;499:485–90.
Thurnher M, Gruenbacher G. T lymphocyte regulation by mevalonate metabolism. Sci Signal. 2015;8:1–10.
Santori FR, Huang P, van de Pavert SA, et al. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. 2015;21:286–98.
Hu X, Wang Y, Hao L-Y, et al. Sterol metabolism controls TH17 differentiation by generating endogenous RORγ agonists. Nat Chem Biol. 2015;11:141–7.
Moon YA. The SCAP/SREBP pathway: a mediator of hepatic steatosis. Endocrinol Metab. 2017;32:6–10.
Chong CR, Xu J, Lu J, Bhat S, Sullivan DJ, Liu JO. Inhibition of angiogenesis by the antifungal drug itraconazole. ACS Chem Biol. 2007;2:263–70.
Liu J, Tian Y, Ding Y, Heng D, Xu K, Liu W, Zhang C. Role of CYP51 in the regulation of T3 and FSH-induced steroidogenesis in female mice. Endocrinology. 2017;158:3974–87.
Van Nistelrooy JGM, Van Den Brink JM, Van Kan JAL, Van Gorcom RFM, De Waard MA. Isolation and molecular characterisation of the gene encoding eburicol 14α-demethylase (CYP51) from Penicillium italicum. Mol Gen Genet. 1996;250:725–33.
Rozman D, Strömstedt M, Tsui LC, Scherer SW, Waterman MR. Structure and mapping of the human lanosterol 14α-demethylase gene (CYP51) encoding the cytochrome p450 involved in cholesterol biosynthesis; comparison of exon/intron organization with other mammalian and fungal CYP genes. Genomics. 1996;38:371–81.
Strömstedt M, Rozman D, Waterman MR. The ubiquitously expressed human CYP51 encodes lanosterol 14α-demethylase, a cytochrome P450 whose expression is regulated by oxysterols. Arch Biochem Biophys. 1996;329:73–81.
Kanda N. Antimycotics suppress interleukin-4 and interleukin-5 production in anti-CD3 plus anti-CD28-stimulated T cells from patients with atopic dermatitis. Japanese J Med Mycol. 2004;45:137–42.
Gong X, Qian H, Zhou X, et al. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola Infection. Cell. 2016;165:1467–78.
Hu M, Yang F, Huang Y, You X, Liu D, Sun S, Sui S-F. Structural insights into the mechanism of human NPC1L1-mediated cholesterol uptake. Sci Adv. 2021;7(29):eabg3188.
Faust PL, Kovacs WJ. Cholesterol biosynthesis and ER stress in peroxisome deficiency. Biochimie. 2014;98:75–85.
Kovacs WJ, Tape KN, Shackelford JE, et al. Peroxisome deficiency causes a complex phenotype because of hepatic SREBP/insig dysregulation associated with endoplasmic reticulum stress. J Biol Chem. 2009;284:7232–45.
Huang B. Song B liang and Xu C: cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab. 2020;2:132–41.
De Giorgi M, Jarrett KE, Burton JC, et al. Depletion of essential isoprenoids and ER stress induction following acute liver-specific deletion of HMG-CoA reductase. J Lipid Res. 2020;61:1675–86.
Nam G-H, Kwon M, Jung H, et al. Statin-mediated inhibition of RAS prenylation activates ER stress to enhance the immunogenicity of KRAS mutant cancer. J Immunother cancer. 2021;9(7):e002474.
Lorbek G, Perše M, Jeruc J, et al. Lessons from hepatocyte-specific Cyp51 knockout mice: impaired cholesterol synthesis leads to oval cell-driven liver injury. Sci Rep. 2015;5:1–11.
Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–27.
Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4:1–25.
Wang L, Zeng X, Ryoo HD, Jasper H. Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet. 2014;10(8):e1004568.
Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. P38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–71.
Xiong G, Hindi SM, Mann AK, et al. The PERK arm of the unfolded protein response regulates satellite cell-mediated skeletal muscle regeneration. Elife. 2017;6:1–27.
Smith M, Wilkinson S. ER homeostasis and autophagy. Essays Biochem. 2017;61:625–35.
Mennerich D, Kellokumpu S, Kietzmann T. Hypoxia and reactive oxygen species as modulators of endoplasmic reticulum and golgi homeostasis. Antioxidants Redox Signal. 2019;30:113–37.
Feelders RA, Newell-Price J, Pivonello R, Nieman LK, Hofland LJ, Lacroix A. Advances in the medical treatment of Cushing’s syndrome. Lancet Diabetes Endocrinol. 2019;7:300–12.
Acknowledgments
The flow cytometry and confocal analysis were performed in Core Facility of Basic Medical Sciences, Shanghai Jiao Tong University School of Medicine. We are grateful for the contributions of their professional assistant. We would like to express our gratitude to EditSprings (https://www.editsprings.cn/) for the expert linguistic services provided.
Funding
This research was funded by the National Natural Science Foundation of China (Grants No. 82001064, 82201086, 82170976, 81970951); Yangfan program of Shanghai Science and Technology Committee (22YF1422100); Fundamental research program funding of Ninth People’s Hospital affiliated to Shanghai Jiao Tong university School of Medicine (JYZZ132); Biological sample bank project of Ninth People’s Hospital Affiliated to Shanghai Jiao Tong university School of Medicine (YBKB202107); The Cross project of Shanghai Ninth people’s hospital affiliated to Shanghai Jiaotong University of Medicine and Shanghai Science and Technology University (JYJC202126); The 15th undergraduate training program for innovation of Shanghai Jiaotong University School of medicine (1521Y591); and the Shanghai Summit & Plateau Disciplines.
Author information
Authors and Affiliations
Contributions
Junhao Yin has a substantial contribution to the conception and design of this study, acquisition of experimental data, analysis of data, formal analysis and draft of the article. Jiayao Fu contributes to interpretation of data and paper writing. Yanxiong Shao contributes to methology. Jiabao Xu contributes to experimental validation and data curation. Hui Li contributes to experimental validation. Changyu Chen contributes to paper writing. Yijie Zhao and Zhanglong Zheng contribute to analysis of data. Chuangqi Yu has a substantial to design of the study. Lingyan Zheng and Baoli Wang have a substantial contribution to revise the article critically for important intellectual content and final approval of the version to be published. All authors read and approved the manuscript for submission.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare no potential conflicts of interest.
Ethical approval
The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Ethics Committee of the Shanghai Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine (Approval sequence: SH9H-2019-T159-2 and SH9H-2021-TK69-1).”
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10238_2022_939_MOESM4_ESM.tif
Additional file 4. Apoptosis evaluation of unactivated CD4+T cells treated with different concentration of KTC.(TIF 1339 KB)
10238_2022_939_MOESM5_ESM.tif
Additional file 5. Long-term treatment of KTC attenuates the symptoms of disease in SS-like NOD/ShiLtj mice. (A) Representative H&E staining of salivary gland tissues of the indicated groups. (B) Lymphocyte infiltration score of the indicated groups. Asterisk indicates P<0.05, double asterisks indicate P<0.01 vs. controlAdditional file 5. Long-term treatment of KTC attenuates the symptoms of disease in SS-like NOD/ShiLtj mice. (A) Representative H&E staining of salivary gland tissues of the indicated groups. (B) Lymphocyte infiltration score of the indicated groups. Asterisk indicates P<0.05, double asterisks indicate P<0.01 vs. control (TIF 3297 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yin, J., Fu, J., Shao, Y. et al. CYP51-mediated cholesterol biosynthesis is required for the proliferation of CD4+ T cells in Sjogren’s syndrome. Clin Exp Med 23, 1691–1711 (2023). https://doi.org/10.1007/s10238-022-00939-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-022-00939-5