Abstract
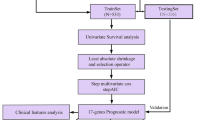
Breast cancer (BC) is the malignancy with the highest mortality rate among women, identification of immune-related biomarkers facilitates precise diagnosis and improvement of the survival rate in early-stage BC patients. 38 hub genes significantly positively correlated with tumor grade were identified based on weighted gene coexpression network analysis (WGCNA) by integrating the clinical traits and transcriptome analysis. Six candidate genes were screened from 38 hub genes basing on least absolute shrinkage and selection operator (LASSO)-Cox and random forest. Four upregulated genes (CDC20, CDCA5, TTK and UBE2C) were identified as biomarkers with the log-rank p < 0.05, in which high expression levels of them showed a poor overall survival (OS) and recurrence-free survival (RFS). A risk model was finally constructed using LASSO-Cox regression coefficients and it possessed superior capability to identify high risk patients and predict OS (p < 0.0001, AUC at 1-, 3- and 5-years are 0.81, 0.73 and 0.79, respectively). Decision curve analysis demonstrated risk score was the best prognostic predictor, and low risk represented a longer survival time and lower tumor grade. Importantly, multiple immune cell types and immunotherapy targets were observed increase in expression levels in high-risk group, most of which were significantly correlated with four genes. In summary, the immune-related biomarkers could accurately predict the prognosis and character the immune responses in BC patients. In addition, the risk model is conducive to the tiered diagnosis and treatment of BC patients.








Similar content being viewed by others
Data availability
All the datasets in this study were downloaded from public databases (GEO, https://www.ncbi.nlm.nih.gov/geo/), and the current research follows the public databases’ access policies and publications guidelines. Users can download relevant data for free, our study is based on open-source data, there are no ethical issues and other conflicts of interest.
References
Bates JP, Derakhshandeh R, Jones L, Webb TJ (2018) Mechanisms of immune evasion in breast cancer. BMC Cancer 18:556. https://doi.org/10.1186/s12885-018-4441-3
Chan CY, Chiu DK, Yuen VW, Law CT, Wong BP, Thu KL, Cescon DW, Soria-Bretones I, Cheu JW, Lee D, Tse AP, Zhang MS, Tan KV, Ng IO, Khong PL, Yau TC, Bray MR, Mak TW, Wong CC (2022) CFI-402257, a TTK inhibitor, effectively suppresses hepatocellular carcinoma. Proc Natl Acad Sci USA 119:e2119514119. https://doi.org/10.1073/pnas.2119514119
Chandler BC, Moubadder L, Ritter CL, Liu M, Cameron M, Wilder-Romans K, Zhang A, Pesch AM, Michmerhuizen AR, Hirsh N, Androsiglio M, Ward T, Olsen E, Niknafs YS, Merajver S, Thomas DG, Brown PH, Lawrence TS, Nyati S, Pierce LJ, Chinnaiyan A, Speers C (2020) TTK inhibition radiosensitizes basal-like breast cancer through impaired homologous recombination. J Clin Invest 130:958–973. https://doi.org/10.1172/JCI130435
Chen K, Xing J, Yu W, Xia Y, Zhang Y, Cheng F, Rao T (2021) Identification and Validation of Hub Genes Associated with Bladder Cancer by Integrated Bioinformatics and Experimental Assays. Front Oncol 11:782981. https://doi.org/10.3389/fonc.2021.782981
Darvin P, Toor SM, Sasidharan Nair V, Elkord E (2018) Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 50:1–11. https://doi.org/10.1038/s12276-018-0191-1
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845
Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau HT, Forero-Torres A, Boccia R, Lippman ME, Somer R, Smakal M, Emens LA, Hrinczenko B, Edenfield W, Gurtler J, von Heydebreck A, Grote HJ, Chin K, Hamilton EP (2018) Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat 167:671–686. https://doi.org/10.1007/s10549-017-4537-5
Dong P, Xiong Y, Yue J, Hanley SJB, Watari H (2018) Tumor-intrinsic PD-L1 signaling in cancer initiation, development and treatment: beyond immune evasion. Front Oncol 8:386. https://doi.org/10.3389/fonc.2018.00386
Farkona S, Diamandis EP, Blasutig IM (2016) Cancer immunotherapy: the beginning of the end of cancer? BMC Med 14:73. https://doi.org/10.1186/s12916-016-0623-5
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14:518–527. https://doi.org/10.1038/nm1764
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1–22
Harbeck N, Gnant M (2017) Breast cancer. Lancet 389:1134–1150. https://doi.org/10.1016/S0140-6736(16)31891-8
Heeke AL, Tan AR (2021) Checkpoint inhibitor therapy for metastatic triple-negative breast cancer. Cancer Metastasis Rev 40:537–547. https://doi.org/10.1007/s10555-021-09972-4
Hu H, Xiang Y, Zhang XY, Deng Y, Wan FJ, Huang Y, Liao XH, Zhang TC (2022) CDCA5 promotes the progression of breast cancer and serves as a potential prognostic biomarker. Oncol Rep 48:172. https://doi.org/10.3892/or.2022.8387
Huang H, Wei Y, Yao H, Chen M, Sun J (2023) Construction of a pancreatic cancer prediction model for oxidative stress-related lncRNA. Funct Integr Genomics 23:118. https://doi.org/10.1007/s10142-023-01048-6
Jiang X, Yuan Y, Tang L, Wang J, Liu Q, Zou X, Duan L (2021) Comprehensive Pan-Cancer Analysis of the Prognostic and Immunological Roles of the METTL3/lncRNA-SNHG1/miRNA-140-3p/UBE2C Axis. Front Cell Dev Biol 9:765772. https://doi.org/10.3389/fcell.2021.765772
Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A, Sica GL, Yu K, Koenig L, Patel NT, Behera M, Wu H, McCausland M, Chen Z, Zhang C, Khuri FR, Owonikoko TK, Ahmed R, Ramalingam SS (2017) Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci USA 114:4993–4998. https://doi.org/10.1073/pnas.1705327114
Kim YJ, Lee G, Han J, Song K, Choi JS, Choi YL, Shin YK (2019) UBE2C overexpression aggravates patient outcome by promoting estrogen-dependent/independent cell proliferation in early hormone receptor-positive and HER2-negative breast cancer. Front Oncol 9:1574. https://doi.org/10.3389/fonc.2019.01574
King JL, Zhang B, Li Y, Li KP, Ni JJ, Saavedra HI, Dong JT (2018) TTK promotes mesenchymal signaling via multiple mechanisms in triple negative breast cancer. Oncogenesis 7:69. https://doi.org/10.1038/s41389-018-0077-z
Kochenderfer JN, Dudley ME, Kassim SH, Somerville RP, Carpenter RO, Stetler-Stevenson M, Yang JC, Phan GQ, Hughes MS, Sherry RM, Raffeld M, Feldman S, Lu L, Li YF, Ngo LT, Goy A, Feldman T, Spaner DE, Wang ML, Chen CC, Kranick SM, Nath A, Nathan DA, Morton KE, Toomey MA, Rosenberg SA (2015) Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J Clin Oncol 33:540–549. https://doi.org/10.1200/JCO.2014.56.2025
Kwapisz D (2021) Pembrolizumab and atezolizumab in triple-negative breast cancer. Cancer Immunol Immunother 70:607–617. https://doi.org/10.1007/s00262-020-02736-z
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. https://doi.org/10.1186/1471-2105-9-559
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28:882–883. https://doi.org/10.1093/bioinformatics/bts034
Lu ZN, Song J, Sun TH, Sun G (2021) UBE2C affects breast cancer proliferation through the AKT/mTOR signaling pathway. Chin Med J (Engl) 134:2465–2474. https://doi.org/10.1097/CM9.0000000000001708
Lu J, Tan J, Yu X (2023) A prognostic model based on tumor microenvironment-related lncRNAs predicts therapy response in pancreatic cancer. Funct Integr Genomics 23:32. https://doi.org/10.1007/s10142-023-00964-x
Lubsen J, Pool J, van der Does E (1978) A practical device for the application of a diagnostic or prognostic function. Methods Inf Med 17:127–129
Maire V, Baldeyron C, Richardson M, Tesson B, Vincent-Salomon A, Gravier E, Marty-Prouvost B, De Koning L, Rigaill G, Dumont A, Gentien D, Barillot E, Roman-Roman S, Depil S, Cruzalegui F, Pierre A, Tucker GC, Dubois T (2013) TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS One 8:e63712. https://doi.org/10.1371/journal.pone.0063712
Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM, Akcakanat A, Chawla A, Curran M, Hwu P, Sharma P, Litton JK, Molldrem JJ, Alatrash G (2014) PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res 2:361–370. https://doi.org/10.1158/2326-6066.CIR-13-0127
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826. https://doi.org/10.1056/NEJMoa041588
Pandya PH, Murray ME, Pollok KE, Renbarger JL (2016) The immune system in cancer pathogenesis: Potential therapeutic approaches. J Immunol Res 2016:4273943. https://doi.org/10.1155/2016/4273943
Qin T, Huang G, Chi L, Sui S, Song C, Li N, Sun S, Li N, Zhang M, Zhao Z, Li L, Li M (2017) Exceptionally high UBE2C expression is a unique phenomenon in basal-like type breast cancer and is regulated by BRCA1. Biomed Pharmacother 95:649–655. https://doi.org/10.1016/j.biopha.2017.08.095
Rawat A, Gopal G, Selvaluxmy G, Rajkumar T (2013) Inhibition of ubiquitin conjugating enzyme UBE2C reduces proliferation and sensitizes breast cancer cells to radiation, doxorubicin, tamoxifen and letrozole. Cell Oncol (Dordr) 36:459–467. https://doi.org/10.1007/s13402-013-0150-8
Vranic S, Cyprian FS, Gatalica Z, Palazzo J (2021) PD-L1 status in breast cancer: Current view and perspectives. Semin Cancer Biol 72:146–154. https://doi.org/10.1016/j.semcancer.2019.12.003
Wang Y, Yao J, Zhu Y, Zhao X, Lv J, Sun F (2022) Knockdown of CDCA5 suppresses malignant progression of breast cancer cells by regulating PDS5A. Mol Med Rep 25:209. https://doi.org/10.3892/mmr.2022.12725
Wang L (2017) Early diagnosis of breast cancer. Sensors (Basel) 17:1572. https://doi.org/10.3390/s17071572
Wimberly H, Brown JR, Schalper K, Haack H, Silver MR, Nixon C, Bossuyt V, Pusztai L, Lannin DR, Rimm DL (2015) PD-L1 expression correlates with tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy in breast cancer. Cancer Immunol Res 3:326–332. https://doi.org/10.1158/2326-6066.CIR-14-0133
Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF, Smylie M, Dummer R, Hill A, Hogg D, Haanen J, Carlino MS, Bechter O, Maio M, Marquez-Rodas I, Guidoboni M, McArthur G, Lebbe C, Ascierto PA, Long GV, Cebon J, Sosman J, Postow MA, Callahan MK, Walker D, Rollin L, Bhore R, Hodi FS, Larkin J (2017) Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 377:1345–1356. https://doi.org/10.1056/NEJMoa1709684
Xiang Z, Li J, Song S, Wang J, Cai W, Hu W, Ji J, Zhu Z, Zang L, Yan R, Yu Y (2019) A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J Exp Clin Cancer Res 38:314. https://doi.org/10.1186/s13046-019-1318-5
Yu GC, Wang LG, Han YY, He QY (2012) clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, Wang Z, Wu Q, Peng H, Wei H, Sun R, Tian Z (2018) Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol 19:723–732. https://doi.org/10.1038/s41590-018-0132-0
Zhang Y, Chen H, Mo H, Hu X, Gao R, Zhao Y, Liu B, Niu L, Sun X, Yu X, Wang Y, Chang Q, Gong T, Guan X, Hu T, Qian T, Xu B, Ma F, Zhang Z, Liu Z (2021) Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 39:1578-1593.e1578. https://doi.org/10.1016/j.ccell.2021.09.010
Zhang S, Wang H, Liu J, Tao T, Zeng Z, Wang M (2022) RGS1 and related genes as potential targets for immunotherapy in cervical cancer: computational biology and experimental validation. J Transl Med 20:334. https://doi.org/10.1186/s12967-022-03526-0
Zheng X, Zhao X, Jin Y, Zhou L, Yang P, Ahmad H, Tian Z (2021) High salt diet contributes to hypertension by weakening the medullary tricarboxylic acid cycle and antioxidant system in Dahl salt-sensitive rats. Biochimie 181:154–161. https://doi.org/10.1016/j.biochi.2020.12.010
Zheng X, Ma H, Wang J, Huang M, Fu D, Qin L, Yin Q (2022) Energy metabolism pathways in breast cancer progression: The reprogramming, crosstalk, and potential therapeutic targets. Transl Oncol 26:101534. https://doi.org/10.1016/j.tranon.2022.101534
Zhu H, Du C, Yuan M, Fu P, He Q, Yang B, Cao J (2020) PD-1/PD-L1 counterattack alliance: multiple strategies for treating triple-negative breast cancer. Drug Discov Today 25:1762–1771. https://doi.org/10.1016/j.drudis.2020.07.006
Zhu Y, Zhu X, Tang C, Guan X, Zhang W (2021) Progress and challenges of immunotherapy in triple-negative breast cancer. Biochim Biophys Acta Rev Cancer 1876:188593. https://doi.org/10.1016/j.bbcan.2021.188593
Funding
This study was financially supported by Fundamental Research Funds for Henan University of Science and Technology (QNY, grant No. 13510001) and A-type Doctoral Talent Project of Henan University of Science and Technology (XWZ, grant No. 13480038).
Author information
Authors and Affiliations
Contributions
XWZ, QNY and WBH planned the research concept and designed it, made provisions for study material. QNY and XWZ collected data and analyzed them, and wrote and approved the manuscript. XWZ and HDM searched for data, wrote programming code, and collected pictures and graphs. JXW provided the help for statistical analysis. YRD, MMF, JXW, JL, MH and AMY helped correct the manuscript. XX provided the valuable comments in revised manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent participate
Not applicable.
Animal ethics
Not applicable.
Consent for publication
Not applicable.
Conflicts of interest
The authors declare that they have no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, X., Ma, H., Dong, Y. et al. Immune-related biomarkers predict the prognosis and immune response of breast cancer based on bioinformatic analysis and machine learning. Funct Integr Genomics 23, 201 (2023). https://doi.org/10.1007/s10142-023-01124-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-01124-x