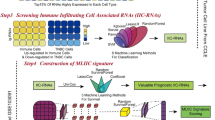
Abstract
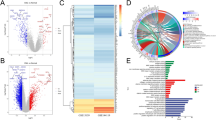
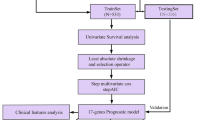
Although medical science has been fully developed, due to the high heterogeneity of triple-negative breast cancer (TNBC), it is still difficult to use reasonable and precise treatment. In this study, based on local optimization-feature screening and genomics screening strategy, we screened 25 feature genes. In multiple machine learning algorithms, feature genes have excellent discriminative diagnostic performance among samples composed of multiple large datasets. After screening at the single-cell level, we identified genes expressed substantially in myeloid cells (MCGs) that have a potential association with TNBC. Based on MCGs, we distinguished two types of TNBC patients who showed considerable differences in survival status and immune-related characteristics. Immune-related gene risk scores (IRGRS) were established, and their validity was verified using validation cohorts. A total of 25 feature genes were obtained, among which CXCL9, CXCL10, CCL7, SPHK1, and TREM1 were identified as the result after single-cell level analysis and screening. According to these entries, the cohort was divided into MCA and MCB subtypes, and the two subtypes had significant differences in survival status and tumor-immune microenvironment. After Lasso-Cox screening, IDO1, GNLY, IRF1, CTLA4, and CXCR6 were selected for constructing IRGRS. There were significant differences in drug sensitivity and immunotherapy sensitivity among high-IRGRS and low-IRGRS groups. We revealed the dynamic relationship between TNBC and TIME, identified a potential biomarker called Granulysin (GNLY) related to immunity, and developed a multi-process machine learning package called “MPMLearning 1.0” in Python.
Graphical Abstract










Similar content being viewed by others
Availability of data and materials
All presented data in this study are available from the corresponding author upon reasonable request.
Change history
13 May 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s10142-024-01370-7
Abbreviations
- TNBC :
-
Triple-negative breast cancer
- ER :
-
Estrogen receptor
- PR :
-
Progesterone receptor
- HER2 :
-
Human epidermal growth factor receptor 2
- TIME :
-
Tumor-immune microenvironment
- TAMs :
-
Tumor-associated macrophages
- ML :
-
Machine learning
- S RA :
-
Sequence read archive
- GEO :
-
Gene expression omnibus
- RF :
-
Random forest
- Oob_score :
-
Out of bag score
- GBDT :
-
Gradient boosting decision tree
- LR :
-
Logistics regression
- SVM :
-
Support vector machine
- TCGA :
-
The Cancer Genome Atlas
- AUC :
-
Area under curve
- ScRNA-seq :
-
Single-cell RNA sequencing
- CXCL9 :
-
C-X-C motif chemokine 9
- CXCL10 :
-
C-X-C motif chemokine 10
- CCL7 :
-
C-C motif chemokine ligand 7
- SPHK1 :
-
Sphingosine kinase 1
- TREM1 :
-
Triggering receptor expressed on myeloid cells 1
- K-M :
-
Kaplan-Meier
- OS :
-
Overall survival
- RFS :
-
Relapse-free survival
- ESTIMATE :
-
Estimation of stromal and immune cells in malignant tumor tissues using expression
- HLA :
-
Human leukocyte antigen
- GSEA :
-
Gene set enrichment analysis
- KEGG :
-
Kyoto Encyclopedia of Genes and Genomes
- NES :
-
Normalized enrichment score
- FDR :
-
False discovery rate
- IRGRS :
-
Immune-related genes risk score
- ROC curve :
-
Receiver operating characteristic curve
- TMB :
-
Tumor mutation burden
- GDSC :
-
Genomics of drug sensitivity in cancer
- CTRP :
-
The Cancer Therapeutics Response Portal
- PRISM :
-
Profiling relative inhibition simultaneously in mixtures
- ICs :
-
Immune checkpoints
- ICIs :
-
Immune checkpoint inhibitors
- PDCD1 :
-
Programmed cell death protein-1
- CD274 :
-
Programmed cell death protein ligand-1
- LAG3 :
-
Lymphocyte activation gene-3
- TIGIT :
-
T cell immunoreceptor with Ig and ITIM domains
- CTLA4 :
-
Cytotoxic T lymphocyte-associated antigen-4
- GNLY :
-
Granulysin
References
Albini A, Sporn MB (2007) The tumour microenvironment as a target for chemoprevention. Nat Rev Cancer 7:139–147. https://doi.org/10.1038/nrc2067
Anderson NR, Minutolo NG, Gill S, Klichinsky M (2021) Macrophage-based approaches for cancer immunotherapymacrophage-based approaches for cancer immunotherapy. Can Res 81:1201–1208. https://doi.org/10.1158/0008-5472.can-20-2990
Bantug GR, Galluzzi L, Kroemer G, Hess C (2018) The spectrum of T cell metabolism in health and disease. Nat Rev Immunol 18:19–34. https://doi.org/10.1038/nri.2017.99
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995. https://doi.org/10.1093/nar/gks1193
Becht E, Giraldo N, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautes-Fridman C, Fridman WH, de Reynies A (2016) Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 17:218. https://doi.org/10.1186/s13059-016-1070-5
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. https://doi.org/10.1038/nbt.4096
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297. https://doi.org/10.1007/BF00994018
Crespo ÂC, Mulik S, Dotiwala F, Ansara JA, Santara SS, Ingersoll K, Ovies C, Junqueira C, Tilburgs T, Strominger JL (2020) Decidual NK cells transfer granulysin to selectively kill bacteria in trophoblasts. Cell 182:1125-1139. e1118. https://doi.org/10.1016/j.cell.2020.07.019
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Fan L, Strasser-Weippl K, Li J-J, St Louis J, Finkelstein DM, Yu K-D, Chen W-Q, Shao Z-M, Goss PE (2014) Breast cancer in China. Lancet Oncol 15:e279–e289. https://doi.org/10.1016/s1470-2045(13)70567-9
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 1189–1232. https://www.jstor.org/stable/2699986
Galluzzi L, Humeau J, Buqué A, Zitvogel L, Kroemer G (2020) Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat Rev Clin Oncol 17:725–741. https://doi.org/10.1038/s41571-020-0413-z
Garrido-Castro AC, Lin NU, Polyak K (2019) Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatmentheterogeneity of triple-negative breast cancer. Cancer Discov 9:176–198. https://doi.org/10.1158/2159-8290.cd-18-1177
Goecks J, Jalili V, Heiser LM, Gray JW (2020) How machine learning will transform biomedicine. Cell 181:92–101. https://doi.org/10.1016/j.cell.2020.03.022
Greener JG, Kandathil SM, Moffat L, Jones DT (2022) A guide to machine learning for biologists. Nat Rev Mol Cell Biol 23:40–55. https://doi.org/10.1038/s41580-021-00407-0
Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21:309–322. https://doi.org/10.1016/j.ccr.2012.02.022
Hedrick CC, Malanchi I (2022) Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol 22:173–187. https://doi.org/10.1038/s41577-021-00571-6
Kharchenko PV (2021) The triumphs and limitations of computational methods for scRNA-seq. Nat Methods 18:723–732. https://doi.org/10.1038/s41592-021-01171-x
LaValley MP (2008) Logistic regression. Circulation 117:2395–2399. https://doi.org/10.1161/circulationaha.106.682658
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS (2017) TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Can Res 77:e108–e110. https://doi.org/10.1158/0008-5472.can-17-0307
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics 27:1739–1740. https://doi.org/10.1093/bioinformatics/btr260
Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H, Wang L, Lu T, Zhang Y, Sun Z (2022) Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat Commun 13:816. https://doi.org/10.1038/s41467-022-28421-6
Maeser D, Gruener RF, Huang RS (2021) oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Briefings in bioinformatics 22:bbab260. https://doi.org/10.1093/bib/bbab260
Mayakonda A, Lin D-C, Assenov Y, Plass C, Koeffler HP (2018) Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 28:1747–1756. https://doi.org/10.1101/gr.239244.118
Myers JA, Miller JS (2021) Exploring the NK cell platform for cancer immunotherapy. Nat Rev Clin Oncol 18:85–100. https://doi.org/10.1038/s41571-020-0426-7
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–457. https://doi.org/10.1038/nmeth.3337
Petegrosso R, Li Z, Kuang RJ (2020) Machine learning and statistical methods for clustering single-cell RNA-sequencing data. Brief Bioinform 21:1209–1223. https://doi.org/10.1093/bib/bbz063
Reina-Campos M, Scharping NE, Goldrath AW (2021) CD8+ T cell metabolism in infection and cancer. Nat Rev Immunol 21:718–738. https://doi.org/10.1038/s41577-021-00537-8
Ren X, Chen X, Zhang X, Jiang S, Zhang T, Li G, Lu Z, Zhang D, Wang S, Qin CJ (2021) Immune microenvironment and response in prostate cancer using large population cohorts. Front Immunol 12:686809. https://doi.org/10.3389/fimmu.2021.686809
Rueda OM, Sammut S-J, Seoane JA, Chin S-F, Caswell-Jin JL, Callari M, Batra R, Pereira B, Bruna A, Ali HR (2019) Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 567:399–404. https://doi.org/10.1038/s41586-019-1007-8
Sharma P, Allison JP (2015) The future of immune checkpoint therapy. Science 348:56–61. https://doi.org/10.1126/science.aaa8172
Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM (2020) B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol 20:294–307. https://doi.org/10.1038/s41577-019-0257-x
Tomczak K, Czerwińska P, Wiznerowicz M (2015) Review the cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol/współczesna Onkologia 2015:68–77. https://doi.org/10.5114/wo.2014.47136
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. https://doi.org/10.1038/nrg2484
Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S (2007) Database resources of the national center for biotechnology information. Nucleic Acids Res 36:D13–D21. https://doi.org/10.1093/nar/gkaa892
Wilkerson MD, Hayes DN (2010) ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26:1572–1573. https://doi.org/10.1093/bioinformatics/btq170
Wruck W, Peuker M, Regenbrecht CR (2014) Data management strategies for multinational large-scale systems biology projects. Brief Bioinform 15:65–78. https://doi.org/10.1093/bib/bbs064
Yang C, Huang X, Li Y, Chen J, Lv Y, Dai S (2021) Prognosis and personalized treatment prediction in TP53-mutant hepatocellular carcinoma: an in silico strategy towards precision oncology. Brief Bioinform 22:bbaa164. https://doi.org/10.1093/bib/bbaa164
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4:1–11. https://doi.org/10.1038/ncomms3612
Zhang J-Y, Wang X-M, Xing X, Xu Z, Zhang C, Song J-W, Fan X, Xia P, Fu J-L, Wang S-Y (2020) Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol 21:1107–1118. https://doi.org/10.1038/s41590-020-0762-x
Acknowledgements
We acknowledge all authors participating in this study for their work and helpful comments.
Funding
This word was supported by the Key Project supported by the Scientific Research Foundation of the Education Bureau of Liaoning Province (2020LZD03); the Liaoning XingLiao Talents Project (XLYC2005014); and the National Natural Science Foundation of China (U1908215). I would like to thank all the teachers and students in the Research Center of Data and Information Science of the School of Medical Devices of Shenyang Pharmaceutical University for their efforts in this project.
Author information
Authors and Affiliations
Contributions
T.L. was responsible for bioinformatics analysis, prepared figures and tables, and designed and wrote the manuscript. S.C., Y.Z., Q.Z., and K.M. carried out data preprocessing. F.Z. also provided some basic code and revised this manuscript. X.J. proofread the manuscript and figures. R.X. and G.L. conceived the concept, instructed bioinformatics analysis, supervised results, and was responsible for its financial supports and the corresponding works.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study is based on public datasets and does not include new data that require ethical approval and consent.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s10142-024-01370-7"
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, T., Chen, S., Zhang, Y. et al. RETRACTED ARTICLE: Ensemble learning-based gene signature and risk model for predicting prognosis of triple-negative breast cancer. Funct Integr Genomics 23, 81 (2023). https://doi.org/10.1007/s10142-023-01009-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-01009-z