Abstract
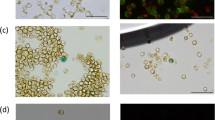
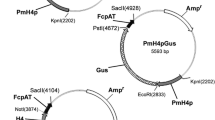
Nitzschia is one of the largest genera of diatoms found in a range of aquatic environments, from freshwater to seawater. This genus contains evolutionarily and ecologically unique species, such as those that have lost photosynthetic capacity or those that live symbiotically in dinoflagellates. Several Nitzschia species have been used as indicators of water pollution. Recently, Nitzschia species have attracted considerable attention in the field of biotechnology. In this study, a transformation method for the marine pennate diatom Nitzschia sp. strain NIES-4635, isolated from the coastal Seto Inland Sea, was established. Plasmids containing the promoter/terminator of the fucoxanthin chlorophyll a/c binding protein gene (fcp, or Lhcf) derived from Nitzschia palea were constructed and introduced into cells by multi-pulse electroporation, resulting in 500 μg/mL nourseothricin-resistant transformants with transformation frequencies of up to 365 colonies per 108 cells. In addition, when transformation was performed using a new plasmid containing a promoter derived from a diatom-infecting virus upstream of the green fluorescent protein gene (gfp), 44% of the nourseothricin-resistant clones exhibited GFP fluorescence. The integration of the genes introduced into the genomes of the transformants was confirmed by Southern blotting. The Nitzschia transformation method established in this study will enable the transformation this species, thus allowing the functional analysis of genes from the genus Nitzschia, which are important species for environmental and biotechnological development.




Similar content being viewed by others
Availability of Data and Materials
The 18S rDNA sequence of strain NIES-4635 is available under DDBJ accession number LC776760. The sequences of N. palea fcp promoter and terminator are available under DDBJ accession numbers LC776935 and LC776936.
References
Angstenberger M, Krischer J, Aktaş O, Büchel C (2019) Knock-down of a ligIV homologue enables DNA integration via homologous recombination in the marine diatom Phaeodactylum tricornutum. ACS Synth Biol 8:57–69
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jézéquel V, Lopez PJ, Lucas S, Mangogna M, Mcginnis K, Medlin LK, Montsant A, Secq M-POL, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, Von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van De Peer Y, Grigoriev IV (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Buhmann MT, Poulsen N, Klemm J, Kennedy MR, Sherrill CD, Kröger N (2014) A tyrosine-rich cell surface protein in the diatom Amphora coffeaeformis identified through transcriptome analysis and genetic transformation. PLoS ONE 9:e110369. https://doi.org/10.1371/journal.pone.0110369
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Conley DJ, Kilham SS, Theriot E (1989) Differences in silica content between marine and freshwater diatoms. Limnol Oceanogr 34:205–212
Costa APT, Schneck F (2022) Diatoms as indicators in running waters: trends of studies on biological assessment and monitoring. Environ Monit Assess 194:695. https://doi.org/10.1007/s10661-022-10383-3
Crowell RM, Nienow JA, Cahoon AB (2019) The complete chloroplast and mitochondrial genomes of the diatom Nitzschia palea (Bacillariophyceae) demonstrate high sequence similarity to the endosymbiont organelles of the dinotom Durinskia baltica. J Phycol 55:352–364
Curran KA, Karim AS, Gupta A, Alper HS (2013) Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab Eng 19:88–97
De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A (2009) Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res 37:e96–e96
Fawzi B, Loudiki M, Oubraim S, Sabour B, Chlaida M (2002) Impact of wastewater effluent on the diatom assemblages structure of a brackish small stream: Oued Hassar (Morocco). Limnologica 32:54–65
Guillard RR, Ryther JH (1962) Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can J Microbiol 8:229–239
Guillory WX, Onyshchenko A, Ruck EC, Parks M, Nakov T, Wickett NJ, Alverson AJ (2018) Recurrent loss, horizontal transfer, and the obscure origins of mitochondrial introns in diatoms (Bacillariophyta). Genome Biol Evol 10:1504–1515
Ifuku K, Yan D, Miyahara M, Inoue-Kashino N, Yamamoto YY, Kashino Y (2015) A stable and efficient nuclear transformation system for the diatom Chaetoceros gracilis. Photosyn Res 123:203–211
Javalkote VS, Pandey AP, Puranik PR, Deshmukh PK (2015) Magnetically responsive siliceous frustules for efficient chemotherapy. Mater Sci Eng C 50:107–116
Johansson ON, Töpel M, Pinder MI, Kourtchenko O, Blomberg A, Godhe A, Clarke AK (2019) Skeletonema marinoi as a new genetic model for marine chain-forming diatoms. Sci Rep 9:5391. https://doi.org/10.1038/s41598-019-41085-5
Kadono T, Tomaru Y, Sato N, Watanabe Y, Suzuki K, Yamada K, Adachi M (2022) Characterization of Chaetoceros lorenzianus-infecting DNA virus-derived promoters of genes from open reading frames of unknown function in Phaeodactylum tricornutum. Mar Genomics 61:100921. https://doi.org/10.1016/j.margen.2021.100921
Kamikawa R, Yubuki N, Yoshida M, Taira M, Nakamura N, Ishida KI, Leander BS, Miyashita H, Hashimoto T, Mayama S, Inagaki Y (2015) Multiple losses of photosynthesis in Nitzschia (Bacillariophyceae). Phycological Res 63:19–28
Kamikawa R, Mochizuki T, Sakamoto M, Tanizawa Y, Nakayama T, Onuma R, Cenci U, Moog D, Speak S, Sarkozi K, Toseland A, Van Oosterhout C, Oyama K, Kato M, Kume K, Kayama M, Azuma T, Ishii KI, Miyashita H, Henrissat B, Lombard V, Win J, Kamoun S, Kashiyama Y, Mayama S, Miyagishima SY, Tanifuji G, Mock T, Nakamura Y (2022) Genome evolution of a nonparasitic secondary heterotroph, the diatom Nitzschia putrida. Sci Adv 8:eabi5075. https://doi.org/10.1126/sciadv.abi5075
Katoh K, Standley DM (2013) MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kira N, Ohnishi K, Miyagawa-Yamaguchi A, Kadono T, Adachi M (2016) Nuclear transformation of the diatom Phaeodactylum tricornutum using PCR-amplified DNA fragments by microparticle bombardment. Mar Genomics 25:49–56
Kröger N, Brunner E (2014) Complex-shaped microbial biominerals for nanotechnology. Wiley Interdiscip Rev Nanomed Nanobiotechnol 6:615–627
Kumazawa M, Nishide H, Nagao R, Inoue-Kashino N, Shen JR, Nakano T, Uchiyama I, Kashino Y, Ifuku K (2022) Molecular phylogeny of fucoxanthin-chlorophyll a/c proteins from Chaetoceros gracilis and Lhcq/Lhcf diversity. Physiol Plant 174:e13598. https://doi.org/10.1111/ppl.13598
Lakshmegowda SB, Rajesh SK, Kandikattu HK, Nallamuthu I, Khanum F (2020) In vitro and in vivo studies on hexane fraction of Nitzschia palea, a freshwater diatom for oxidative damage protective and anti-inflammatory response. Rev Bras Farmacogn 30:189–201
Laviale M, Beaussart A, Allen J, Quilès F, El-Kirat-Chatel S (2019) Probing the adhesion of the common freshwater diatom Nitzschia palea at nanoscale. ACS Appl Mater Interfaces 11:48574–48582
Mann DG (1986) Methods of sexual reproduction in Nitzschia: systematic and evolutionary implications. Diatom Res 1:193–203
Mann DG, Trobajo R, Sato S, Li C, Witkowski A, Rimet F, Ashworth MP, Hollands RM, Theriot EC (2021) Ripe for reassessment: a synthesis of available molecular data for the speciose diatom family Bacillariaceae. Mol Phylogenet Evol 158:106985. https://doi.org/10.1016/j.ympev.2020.106985
Mann DG, Yamada N, Bolton JJ, Witkowski A, Trobajo R (2023) Nitzschia captiva sp. nov. (Bacillariophyta), the essential prey diatom of the kleptoplastic dinoflagellate Durinskia capensis, compared with N. agnita. N Kuetzingioides and Other Species Phycologia 62:136–151
Miyagawa A, Okami T, Kira N, Yamaguchi H, Ohnishi K, Adachi M (2009) Research note: high efficiency transformation of the diatom Phaeodactylum tricornutum with a promoter from the diatom Cylindrotheca fusiformis. Phycological Res 57:142–146
Miyagawa-Yamaguchi A, Okami T, Kira N, Yamaguchi H, Ohnishi K, Adachi M (2011) Stable nuclear transformation of the diatom Chaetoceros sp. Phycological Res 59:113–119
Miyahara M, Aoi M, Inoue-Kashino N, Kashino Y, Ifuku K (2013) Highly efficient transformation of the diatom Phaeodactylum tricornutum by multi-pulse electroporation. Biosci Biotechnol Biochem 77:874–876
Muto M, Fukuda Y, Nemoto M, Yoshino T, Matsunaga T, Tanaka T (2013) Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580—a high triglyceride producer. Mar Biotechnol 15:48–55
Naser I, Yabu Y, Maeda Y, Tanaka T (2022) Highly efficient genetic transformation methods for the marine oleaginous diatom Fistulifera solaris. Mar Biotechnol. https://doi.org/10.1007/s10126-022-10189-x
Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B (1995) Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycles 9:359–372
Nemoto M, Iwaki S, Moriya H, Monden Y, Tamura T, Inagaki K, Mayama S, Obuse K (2020) Comparative gene analysis focused on silica cell wall formation: identification of diatom-specific SET domain protein methyltransferases. Mar Biotechnol 22:551–563
Oliver A, Podell S, Pinowska A, Traller JC, Smith SR, Mcclure R, Beliaev A, Bohutskyi P, Hill EA, Rabines A, Zheng H, Allen LZ, Kuo A, Grigoriev IV, Allen AE, Hazlebeck D, Allen EE (2021) Diploid genomic architecture of Nitzschia inconspicua, an elite biomass production diatom. Sci Rep 11:15592. https://doi.org/10.1038/s41598-021-95106-3
Poulsen N, Chesley PM, Kröger N (2006) Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae)1. J Phycol 42:1059–1065
Ramachandra TV, Mahapatra DM, Karthick B, Gordon R (2009) Milking diatoms for sustainable energy: biochemical engineering versus gasoline-secreting diatom solar panels. Ind Eng Chem Res 48:8769–8788
Round FE, Crawford RM, Mann DG (1990) The diatoms: biology and morphology of the genera. Cambridge University Press, Cambridge
Rovira L, Trobajo R, Sato S, Ibáñez C, Mann DG (2015) Genetic and physiological diversity in the diatom Nitzschia inconspicua. J Eukaryot Microbiol 62:815–832
Sanniyasi E, Patrick APR, Rajagopalan K, Gopal RK, Damodharan R (2022) Characterization and in vitro anticancer potential of exopolysaccharide extracted from a freshwater diatom Nitzschia palea (Kütz.) W.Sm. 1856. Sci Rep 12:22114. https://doi.org/10.1038/s41598-022-24662-z
Shin YJ, Lim JM, Park JH, Choi DW, Hwang MS, Park EJ, Min SR, Kim SW, Jeong WJ (2016) Characterization of PyGUS gene silencing in the red macroalga, Pyropia yezoensis. Plant Biotechnol Rep 10:359–367
Sprecher BN, Buck JM, Ropella LL, Ramsperger A, Kroth PG, Yamada N (2023) Genetic transformation methods for diatom Nitzschia captiva: New tools to better understand dinotom endosymbiosis. Algal Res 72:103136. https://doi.org/10.1016/j.algal.2023.103136
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Tanaka T, Maeda Y, Veluchamy A, Tanaka M, Abida H, Maréchal E, Bowler C, Muto M, Sunaga Y, Tanaka M, Yoshino T, Taniguchi T, Fukuda Y, Nemoto M, Matsumoto M, Wong PS, Aburatani S, Fujibuchi W (2015) Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 27:162–176
Theriot EC, Ashworth M, Ruck E, Nakov T, Jansen RK (2010) A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Plant Ecol Evol 143:278–296
Trobajo R, Mann DG, Chepurnov VA, Clavero E, Cox EJ (2006) Taxonomy, life cycle, and auxosporulation of Nitzschia fonticola (Bacillariophyta)1. J Phycol 42:1353–1372
Tsuchiya T, Kameya N, Nakamura I (2009) Straight Walk: a modified method of ligation-mediated genome walking for plant species with large genomes. Anal Biochem 388:158–160
Wang Y, Mu W, Sun X, Lu X, Fan Y, Liu Y (2020) Physiological response and removal ability of freshwater diatom Nitzschia palea to two organophosphorus pesticides. Chem Ecol 36:881–902
Watanabe MM and Satake KN (1991) NIES-collection LIST of STRAINS third edition, microalgae and protozoa. National Institute for Environmental Studies 31–32
Wyness AJ, Fortune I, Blight AJ, Browne P, Hartley M, Holden M, Paterson DM (2021) Ecosystem engineers drive differing microbial community composition in intertidal estuarine sediments. PLoS ONE 16:e0240952. https://doi.org/10.1371/journal.pone.0240952
Xing RL, Ma WW, Shao YW, Cao XB, Su C, Song HX, Su Q, Zhou GF (2018) Growth and potential purification ability of Nitzschia sp. benthic diatoms in sea cucumber aquaculture wastewater. Aquac Res 49:2644–2652
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33:103–119
Yi Z, Xu M, Di X, Brynjolfsson S, Fu W (2017) Exploring valuable lipids in diatoms. Front Mar Sci 4. https://doi.org/10.3389/fmars.2017.00017
Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE (2000) Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol 36:379–386
Acknowledgements
We thank Dr. Wataru Sakamoto at the Institute of Plant Science and Resources, Okayama University, Japan, for his assistance with the microparticle bombardment. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Funding
This study was supported by the Program to Disseminate Tenure Tracking System from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan; Grants-in-Aid for Scientific Research (C) (No. 18K05818); and the Asahi Glass Foundation to MN.
Author information
Authors and Affiliations
Contributions
Michiko Nemoto designed the research; Koki Okada, Yu Morimoto, and Yukine Shiraishi performed the experiments; Koki Okada, Yu Morimoto, Yukine Shiraishi, Takashi Tamura, Shigeki Mayama, Takashi Kadono, Masao Adachi, Kentaro Ifuku and Michiko Nemoto analyzed the data; Koki Okada, Takashi Kadono, Masao Adachi, Kentaro Ifuku, and Michiko Nemoto wrote and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Competing Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Okada, K., Morimoto, Y., Shiraishi, Y. et al. Nuclear Transformation of the Marine Pennate Diatom Nitzschia sp. Strain NIES-4635 by Multi-Pulse Electroporation. Mar Biotechnol 25, 1208–1219 (2023). https://doi.org/10.1007/s10126-023-10273-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-023-10273-w