Abstract
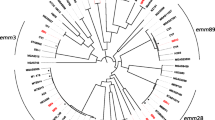
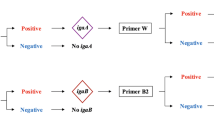
Haemophilus influenzae is a common cause of mucosal infections that warrants accurate surveillance. We aimed to assess the prevalence of the species in clinical specimens, and characterise population structure and resistance to aminopenicillins by whole genome sequencing.We assessed the point prevalence by entering the database records of 1 day in Denmark and examined the genome sequences of nationwide, collected isolates from the same day. The prevalence of H. influenzae in clinical samples on the 10th of January 2018 was 1.78 per 100,000 person-days (all samples), and 2.47 per 1000 hospital bed-days (hospital samples). Of 2009 bacteria deemed clinically relevant and collected in a concerted action by the Danish departments of clinical microbiology, 62 (3.1%) were H. influenzae. All 62 isolates belonged to phylogenetic group I and were unencapsulated. Three strains from separate Danish regions had identical core genome sequences, but a small number of intergenic mutations testified to circulating clones, rather than individual cases of patient-to-patient transmission. The TEM-1 β-lactamase gene was present in 24 strains, while 13 strains were genetically categorised as ampicillin-resistant due to substitutions in penicillin-binding protein 3; shared patterns of amino acid substitutions in unrelated strains indicated putative lateral transfer of chromosomal resistance. Circulating clones of H. influenzae are frequent, and host factors, rather than direct transmission of epidemic strains, may be the primary cause of infection. The bleak presence of ampicillin resistance revealed by sequencing of point prevalence strains underscores the necessity for close examination of testing methods.




Similar content being viewed by others
Data availability
Accession numbers are given in the “Materials and methods” section. The dataset analysed as part of the current study is available from the corresponding author on reasonable request.
References
Van Eldere J, Slack MPE, Ladhani S, Cripps AW (2014) Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis 14:1281–1292. https://doi.org/10.1016/S1473-3099(14)70734-0
Slack MPE (2015) A review of the role of Haemophilus influenzae in community-acquired pneumonia. Pneumonia 6:26–43. https://doi.org/10.15172/pneu.2015.6/520
Heliodoro CIM, Bettencourt CR, Bajanca-Lavado MP, Portuguese Group for the Study of Haemophilus influenzae invasive infection (2020) Molecular epidemiology of invasive Haemophilus influenzae disease in Portugal: an update of the post-vaccine period, 2011-2018. Eur J Clin Microbiol Infect Dis 39:1471–1480. https://doi.org/10.1007/s10096-020-03865-0
Gessner BD, Adegbola RA (2008) The impact of vaccines on pneumonia: key lessons from Haemophilus influenzae type b conjugate vaccines. Vaccine 26:B3–B8. https://doi.org/10.1016/j.vaccine.2008.04.013
Ulanova M (2020) Invasive Haemophilus influenzae serotype a disease in the Hib conjugate vaccine era: where are we going? Clin Infect Dis. https://doi.org/10.1093/cid/ciaa868 Online ahead of print
Gallo MC, Kirkham C, Eng S, Bebawee RS, Kong Y, Pettigrew MM, Tettelin H, Murphy TF (2018) Changes in IgA protease expression are conferred by changes in genomes during persistent infection by nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease. Infect Immun 86:e00313–e00318. https://doi.org/10.1128/IAI.00313-18
Nelson JD (1974) Editorial: Should ampicillin be abandoned for treatment of Haemophilus influenzae disease? JAMA 229:322–324
Elwell LP, De Graaff J, Seibert D, Falkow S (1975) Plasmid-linked ampicillin resistance in Haemophilus influenzae type b. Infect Immun 12:404–410. https://doi.org/10.1128/IAI.12.2.404-410.1975
Søndergaard A, Lund M, Nørskov-Lauritsen N (2015) TEM-1-encoding small plasmids impose dissimilar fitness costs on Haemophilus influenzae and Haemophilus parainfluenzae. Microbiology 161:2310–2315. https://doi.org/10.1099/mic.0.000183
Trepod CM, Mott JE (2005) Elucidation of essential and nonessential genes in the Haemophilus influenzae Rd cell wall biosynthetic pathway by targeted gene disruption. Antimicrob Agents Chemother 49:824–826. https://doi.org/10.1128/AAC.49.2.824-826.2005
Ubukata K, Shibasaki Y, Yamamoto K, Chiba N, Hasegawa K, Takeuchi Y, Sunakawa K, Inoue M, Konno M (2001) Association of amino acid substitutions in penicillin-binding protein 3 with β-lactam resistance in β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob Agents Chemother 45:1693–1699. https://doi.org/10.1128/AAC.45.6.1693-1699.2001
Skaare D, Anthonisen IL, Kahlmeter G, Matuschek E, Natås OB, Steinbakk M, Sundsfjord A, Kristiansen BE (2014) Emergence of clonally related multidrug resistant Haemophilus influenzae with penicillin-binding protein 3-mediated resistance to extended-spectrum cephalosporins, Norway, 2006 to 2013. Euro Surveill 19(49):20986. https://doi.org/10.2807/1560-7917.es2014.19.49.20986
García-Cobos S, Campos J, Lázaro E, Román F, Cercenado E, García-Rey C, Pérez-Vázquez M, Oteo J, de Abajo F (2007) Ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob Agents Chemother 51:2564–2573. https://doi.org/10.1128/AAC.00354-07
Deghmane AE, Hong E, Chehboub S, Terrade A, Falguières M, Sort M, Harrison O, Jolley KA, Taha MK (2019) High diversity of invasive Haemophilus influenzae isolates in France and the emergence of resistance to third generation cephalosporins by alteration of ftsI gene. J Inf Secur 79:7–14. https://doi.org/10.1016/j.jinf.2019.05.007
Hotomi M, Fujihara K, Billal DS, Suzuki K, Nishimura T, Baba S, Yamanaka N (2007) Genetic characteristics and clonal dissemination of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae strains isolated from the upper respiratory tract of patients in Japan. Antimicrob Agents Chemother 51:3969–3976. https://doi.org/10.1128/AAC.00422-07
Han MS, Jung HJ, Lee HJ, Choi EH (2019) Increasing prevalence of group III penicillin-binding protein 3 mutations conferring high-level resistance to beta-lactams among nontypeable Haemophilus influenzae isolates from children in Korea. Microb Drug Resist 25:567–576. https://doi.org/10.1089/mdr.2018.0342
WHO Global priority list of antibiotic-resistant bacteria 2017. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf
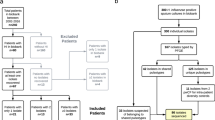
Rebelo AR, Ibfelt T, Bortolaia V, Leekitcharoenphon P, Hansen DS, Nielsen HL, Ellermann-Eriksen S, Kemp M, Røder BL, Frimodt-Møller N, Søndergaard TS, Coia JE, Østergaard C, Pedersen M, Westh H, Aarestrup FM () One day in Denmark: nationwide point-prevalence survey of human bacterial isolates and comparison of classical and whole-genome sequence-based species identification methods (submitted)
Voldstedlund M, Haarh M, Mølbak K, MiBa Board of Representatives (2014) The Danish Microbiology Database (MiBa) 2010 to 2013. Euro Surveill 19:20667. https://doi.org/10.2807/1560-7917.es2014.19.1.20667
Seemann T, Goncalves da Silva A, Bulach DM, Schultz MB, Kwong JC, Howden BP. Nullarbor Github https://github.com/tseemann/nullarbor
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Souvorov A, Agarwala R, Lipman DJ (2018) SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol 19:153. https://doi.org/10.1186/s13059-018-1540-z
Nedergaard S, Kobel CM, Nielsen MB, Møller RT, Jensen AB, Nørskov-Lauritsen N (2019) Whole genome sequencing of Aggregatibacter actinomycetemcomitans cultured from blood stream infections reveals three major phylogenetic groups including a novel lineage expressing serotype a membrane O polysaccharide. Pathogens 8:256. https://doi.org/10.3390/pathogens8040256
Wattam AR, Brettin T, Davis JJ, Gerdes S, Kenyon R, Machi D, Mao C, Olson R, Overbeek R, Pusch GD, Shukla MP, Stevens R, Vonstein V, Warren A, Xia F, Yoo H (2018) Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol Biol 1704:79–101. https://doi.org/10.1007/978-1-4939-7463-4_4
Sinha S, Mell JC, Redfield RJ (2012) Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J Bacteriol 194:5245–5254. https://doi.org/10.1128/JB.00671-12
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Søndergaard A, San Millan A, Santos-Lopez A, Nielsen SM, Gonzalez-Zorn B, Nørskov-Lauritsen N (2012) Molecular organization of small plasmids bearing blaTEM-1 and conferring resistance to β-lactams in Haemophilus influenzae. Antimicrob Agents Chemother 56:4958–4960. https://doi.org/10.1128/AAC.00408-12
Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG (2003) Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. https://doi.org/10.1128/jcm.41.4.1623-1636.2003
De Chiara M, Hood D, Muzzi A, Pickard DJ, Perkins T, Pizza M, Dougan G, Rappuoli R, Moxon ER, Soriani M, Donati C (2014) Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure. Proc Natl Acad Sci U S A 111:5439–44. https://doi.org/10.1073/pnas.1403353111
Rajendra KC, Leong KWC, Harkness NM, Lachowicz J, Gautam SS, Cooley LA, McEwan B, Petrovski S, Karupiah G, O'Toole RF. Whole-genome analyses reveal gene content differences between nontypeable Haemophilus influenzae isolates from chronic obstructive pulmonary disease compared to other clinical phenotypes. Microb Genom 2020:6. https://doi.org/10.1099/mgen.0.000405
Hansen KH, Andreasen MR, Pedersen MS, Westh H, Jelsbak L, Schønning K (2019) Resistance to piperacillin/tazobactam in Escherichia coli resulting from extensive IS26-associated gene amplification of blaTEM-1. J Antimicrob Chemother 74:3179–3183. https://doi.org/10.1093/jac/dkz349
Skaare D, Anthonisen IL, Caugant DA, Jenkins A, Steinbakk M, Strand L, Sundsfjord A, Tveten Y, Kristiansen BE (2014) Multilocus sequence typing and ftsI sequencing: a powerful tool for surveillance of penicillin-binding protein 3-mediated beta-lactam resistance in nontypeable Haemophilus influenzae. BMC Microbiol 14:131. https://doi.org/10.1186/1471-2180-14-131
Honda H, Sato T, Shinagawa M, Fukushima Y, Nakajima C, Suzuki Y, Kuronuma K, Takahashi S, Takahashi H, Yokota S-I (2020) In vitro derivation of fluoroquinolone-resistant mutants from multiple lineages of Haemophilus influenzae and identification of mutations associated with fluoroquinolone resistance. Antimicrob Agents Chemother 64:e01500–e01519. https://doi.org/10.1128/AAC.01500-19
Nørskov-Lauritsen N (2014) Classification, identification, and clinical significance of Haemophilus and Aggregatibacter species with host specificity for humans. Clin Microbiol Rev 27:214–240. https://doi.org/10.1128/CMR.00103-13
Harris TM, Price EP, Sarovich DS, Nørskov-Lauritsen N, Beissbarth J, Chang AB, Smith-Vaughan HC (2020) Comparative genomic analysis identifies X-factor (haemin)-independent Haemophilus haemolyticus: a formal re-classification of ‘Haemophilus intermedius’. Microb Genom 6:e000303. https://doi.org/10.1099/mgen.0.000303
Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755. https://doi.org/10.1086/647952
Barry AL, Fuchs PC, Brown SD (2001) Identification of beta-lactamase-negative, ampicillin-resistant strains of Haemophilus influenzae with four methods and eight media. Antimicrob Agents Chemother 45:1585–1588. https://doi.org/10.1128/AAC.45.5.1585-1588.2001
Skaare D, Lia A, Hannisdal A, Tveten Y, Matuschek E, Kahlmeter G, Kristiansen BE (2015) Haemophilus influenzae with non-beta-lactamase-mediated beta-lactam resistance: easy to find but hard to categorize. J Clin Microbiol 53:3589–3595. https://doi.org/10.1128/JCM.01630-15
Nørskov-Lauritsen N, Ridderberg W, Erikstrup LT, Fuursted K (2011) Evaluation of disk diffusion methods to detect low-level β-lactamase-negative ampicillin-resistant Haemophilus influenzae. APMIS 119:385–392. https://doi.org/10.1111/j.1600-0463.2011.02745.x
Maughan H, Redfield RJ (2009) Extensive variation in natural competence in Haemophilus influenzae. Evolution 63(7):1852–1866. https://doi.org/10.1111/j.1558-5646.2009.00658.x
Acknowledgements
Members of the ODiD Consortium are as follows: Ana Rita Rebelo, Valeria Bortolaia, Pimlapas Leekitcharoenphon, and Frank Møller Aarestrup (Technical University of Denmark); Hans Linde Nielsen (DCM Aalborg); Svend Ellermann-Eriksen (DCM Aarhus); Turid Snekloth Søndergaard (DCM Sønderborg); John Eugenio Coia (DCM Esbjerg); Claus Østergaard (DCM Vejle); Michael Kemp (University of Southern Denmark); Bent Løwe Røder (DCM Slagelse); Niels Frimodt-Møller (DCM Rigshospitalet); Dennis Schrøder Hansen (DCM Herlev); Henrik Westh (DCM Hvidovre).
Author information
Authors and Affiliations
Consortia
Contributions
The study was conceived and planned by NNL. JL retrieved data from MiBa, and CMK performed the major part of the bioinformatics analysis. NNL analysed data and wrote the first draft of the manuscript, which was revised critically for clinical content by NP, HLN, and DSH. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval and informed consent
Not applicable to the present study. Reported culture of H. influenzae and collected bacterial strains are linked to specimen type and a regional DCM. Information cannot be linked to patient records or original sample number, and according to Danish law, approval from science ethics committees is not required.
Conflict of interest
The authors declare no competing interests.
Disclaimer
The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of individual DCMs, the Danish Society of Clinical Microbiology, or the Danish Microbiology Database.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nørskov-Lauritsen, N., Pedersen, N., Lam, J.U.H. et al. Haemophilus influenzae one day in Denmark: prevalence, circulating clones, and dismal resistance to aminopenicillins. Eur J Clin Microbiol Infect Dis 40, 2077–2085 (2021). https://doi.org/10.1007/s10096-021-04247-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04247-w