Abstract
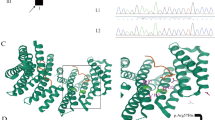
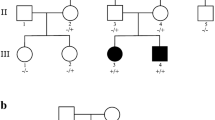
Human exocyst complex is an evolutionary conserved multimeric complex composed of proteins encoded by eight genes EXOC1-EXOC8. It is known that the exocyst complex plays a role in ciliogenesis, cytokinesis, cell migration, autophagy, and fusion of secretory vesicles. Recently, loss of function variants in EXOC7 and EXOC8 has been associated with abnormalities of cerebral cortical development leading to a neurodevelopmental phenotype. Neurodevelopmental disorders are a huge group of clinically and genetically heterogeneous disorders. In the present study, we recruited a large consanguineous family segregating a neurodevelopmental disorder in an autosomal recessive form. We performed clinical phenotyping by imaging the patient’s brain followed by whole exome sequencing examining DNA from two affected individuals. The clinical phenotypes of the disease were suggestive of brain atrophy. Clinical examination revealed intellectual impairment with hypertonia and brisk reflexes. WES followed by Sanger sequencing revealed a novel homozygous nonsense mutation [EXOC8; NM_175876.5; c.1714G > T; p.(Glu572Ter)] in the DNA of affected individuals. Both parents of the patients were heterozygous for the identified mutation. All the pathogenicity prediction softwares predicted the identified variant as disease causing. This study reports a second protein-truncating variant in EXOC8. The findings confirm that loss of function variants in EXOC8 underlies a neurodevelopmental disorder. The identification of a protein-truncating variant in EXOC8 in the current study can be helpful in establishing genotype–phenotype correlations. Our results also provide new insights into genetic counseling and clinical management for the affected individuals.


Similar content being viewed by others
References
Mei K, Li Y, Wang S et al (2018) Cryo-EM structure of the exocyst complex. Nat Struct Mol Biol 25(2):139–146. https://doi.org/10.1038/s41594-017-0016-2
Heider MR, Munson M (2012) Exorcising the exocyst complex. Traffic 13:898–907. https://doi.org/10.1111/j.1600-0854.2012.01353.x
Luo G, Zhang J, Luca FC, Guo W (2013) Mitotic phosphorylation of Exo84 disrupts exocyst assembly and arrests cell growth. J Cell Biol 202:97–111. https://doi.org/10.1083/jcb.201211093
Wu B, Guo W (2015) The exocyst at a glance. J Cell Sci 128:2957–2964. https://doi.org/10.1242/jcs.156398
He B, Guo W (2009) The exocyst complex in polarized exocytosis. Curr Opin Cell Biol 21:537–542. https://doi.org/10.1016/j.ceb.2009.04.007 (Epub 2009 May 25)
Dixon-Salazar TJ, Silhavy JL, Udpa N et al (2012) Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med 4(138):13878. https://doi.org/10.1126/scitranslmed.3003544
Coulter ME, Musaev D, DeGennaro EM et al (2020) Regulation of human cerebral cortical development by EXOC7 and EXOC8, components of the exocyst complex, and roles in neural progenitor cell proliferation and survival. Genet Med 22:1040–1050. https://doi.org/10.1038/s41436-020-0758-9
Banerjee TD, Middleton F, Faraone SV (2007) Environmental risk factors for attention-deficit hyperactivity disorder. Acta Paediatr 96(9):1269–1274. https://doi.org/10.1111/j.1651-2227.2007.00430.x
Khan S, Lin S, Harlalka GV et al (2019) BBS5 and INPP5E mutations associated with ciliopathy disorders in families from Pakistan. Ann Hum Genet 83(6):477–482. https://doi.org/10.1111/ahg.12336
Bibi F, Haider N, Din SU et al. (2020) Sequence variants in three genes underlying leukodystrophy in Pakistani families. Int J Dev Neurosci 80(5):380–388. https://doi.org/10.1002/jdn.10036
Cardoso AR, Lopes-Marques M, Silva RM et al (2019) Essential genetic findings in neurodevelopmental disorders. Hum Genomics 13:31. https://doi.org/10.1186/s40246-019-0216-4
Reuter MS, Tawamie H, Buchert R et al (2017) Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiat 74(3):293–299. https://doi.org/10.1001/jamapsychiatry.2016.3798
Parenti I, Rabaneda LG, Schoen H, Novarino G (2020) Neurodevelopmental disorders: from genetics to functional pathways. Trends Neurosci 43(8):608–621. https://doi.org/10.1016/j.tins.2020.05.004
Umair M, Ahmad F, Ullah A (2018) Whole exome sequencing as a diagnostic tool for genetic disorders in Pakistan. Pak J Med Res 57(2):90–91
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual; preparation and analysis of eukaryotic genomic DNA, 3rd edn. Cold Spring Harbor Laboratory Press, New York, p 2001
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
McKenna A, Hanna M, Banks E (2010) The genome analysis toolkit: a MapReduce framework for analyzing next generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT (2005) Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J 24(12):2064–2074. https://doi.org/10.1038/sj.emboj.7600699
Strande NT, Riggs ER, Buchanan AH et al (2017) Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the clinical genome resource. Am J Hum Genet 100(6):895–906. https://doi.org/10.1016/j.ajhg.2017.04.015
Dickinson ME, Flenniken AM, Ji X et al (2016) High-throughput discovery of novel developmental phenotypes. Nature 537:508–514. https://doi.org/10.1038/nature19356
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ullah, A., Krishin, J., Haider, N. et al. A novel nonsense variant in EXOC8 underlies a neurodevelopmental disorder. Neurogenetics 23, 203–212 (2022). https://doi.org/10.1007/s10048-022-00692-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-022-00692-7