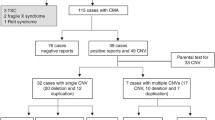
Abstract
Copy number variations (CNVs) are highly implicated in the etiology of neurodevelopmental disorders (NDDs), and chromosomal microarray analysis (CMA) has been recommended as a first-tier test for many NDDs. We undertook a study to identify clinically relevant CNVs and genes in an ethnically homogenous population of the United Arab Emirates. We genotyped 98 patients with NDDs using genome-wide chromosomal microarray analysis, and observed 47.1% deletion and 52.9% duplication CNVs, of which 11.8% are pathogenic, 23.5% are likely pathogenic, and 64.7% VOUS. The average size of copy number losses (3.9 Mb) was generally higher than of gains (738.4 kb). Analysis of VOUS CNVs for constrained genes (enrichment for brain critical exons and high pLI genes) yielded 7 unique genes. Among these 7 constrained genes, we propose FNTA and PXK as potential candidate genes for neurodevelopmental disorders, which warrants further investigation. Thirty-two overlapping CNVs (Decipher and ClinVar) containing the FNTA gene were previously identified in NDD patients and 6 overlapping CNVs (Decipher and ClinVar) containing the PXK gene were previously identified in NDD patients. Our study supports the utility of CMA for CNV profiling which aids in precise genetic diagnosis and its integration into therapeutics and management of NDD patients.




Similar content being viewed by others
Data availability
All data are presented in the manuscript and supplementary information.
Code availability
NA.
References
Hansen BH, Oerbeck B, Skirbekk B, Petrovski BE, Kristensen H (2018) Neurodevelopmental disorders: prevalence and comorbidity in children referred to mental health services. Nord J Psychiatry 72(4):285–291. https://doi.org/10.1080/08039488.2018.1444087
Parenti I, Rabaneda LG, Schoen H, Novarino G (2020) Neurodevelopmental disorders: from genetics to functional pathways. Trends Neurosci 43(8):608–621. https://doi.org/10.1016/j.tins.2020.05.004
Gilissen C et al (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature 511(7509):344–347. https://doi.org/10.1038/nature13394
First MB (2013) Diagnostic and statistical manual of mental disorders, 5th edition, and clinical utility. J Nerv Ment Dis 201(9):727–9. https://doi.org/10.1097/NMD.0b013e3182a2168a
M. Woodbury-Smith et al. (2017) Variable phenotype expression in a family segregating microdeletions of the NRXN1 and MBD5 autism spectrum disorder susceptibility genes. NPJ Genom Med. 2. https://doi.org/10.1038/s41525-017-0020-9
Woodbury-Smith M et al (2017) Mutations in RAB39B in individuals with intellectual disability, autism spectrum disorder, and macrocephaly. Mol Autism 8:59. https://doi.org/10.1186/s13229-017-0175-3
M. Woodbury-Smith et al (2022) Mutational landscape of autism spectrum disorder brain tissue. Genes. 13 (2);207 [Online]. Available: https://www.mdpi.com/2073-4425/13/2/207
Girirajan S, Campbell CD, Eichler EE (2011) Human copy number variation and complex genetic disease. Annu Rev Genet 45:203–226. https://doi.org/10.1146/annurev-genet-102209-163544
Jang W et al (2019) Chromosomal microarray analysis as a first-tier clinical diagnostic test in patients with developmental delay/intellectual disability, autism spectrum disorders, and multiple congenital anomalies: a prospective multicenter study in Korea. Ann Lab Med 39(3):299–310. https://doi.org/10.3343/alm.2019.39.3.299
Tammimies K et al (2015) Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA 314(9):895–903. https://doi.org/10.1001/jama.2015.10078
al-Gazali LI, Bener A, Abdulrazzaq YM, Micallef R, al-Khayat AI, Gaber T (1997) Consanguineous marriages in the United Arab Emirates. J Biosoc Sci. 29(4):491–7. https://doi.org/10.1017/s0021932097004914
Akter H et al (2021) Whole exome sequencing uncovered highly penetrant recessive mutations for a spectrum of rare genetic pediatric diseases in Bangladesh. NPJ Genom Med. 6(1):14. https://doi.org/10.1038/s41525-021-00173-0
Richards S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Subramanian A et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Uddin M et al (2014) Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet 46(7):742–747. https://doi.org/10.1038/ng.2980
Lek M et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. https://doi.org/10.1038/nature19057
Sunkin SM et al (2013) Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 41(Database issue):D996–D1008. https://doi.org/10.1093/nar/gks1042
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628. https://doi.org/10.1038/nmeth.1226
Nassir N et al (2021) Single-cell transcriptome identifies molecular subtype of autism spectrum disorder impacted by de novo loss-of-function variants regulating glial cells. Hum Genomics 15(1):68. https://doi.org/10.1186/s40246-021-00368-7
Cao J et al (2020) A human cell atlas of fetal gene expression. Science. 370 (6518). https://doi.org/10.1126/science.aba7721
Szklarczyk D et al (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(Database issue):D447-52. https://doi.org/10.1093/nar/gku1003
Pinner AL, Mueller TM, Alganem K, McCullumsmith R, Meador-Woodruff JH (2020) Protein expression of prenyltransferase subunits in postmortem schizophrenia dorsolateral prefrontal cortex. Transl Psychiatry 10(1):3. https://doi.org/10.1038/s41398-019-0610-7
Ruderfer DM et al (2016) Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat Genet 48(10):1107–1111. https://doi.org/10.1038/ng.3638
Jones KL et al (2017) Autism with intellectual disability is associated with increased levels of maternal cytokines and chemokines during gestation. Mol Psychiatry 22(2):273–279. https://doi.org/10.1038/mp.2016.77
Szczaluba K et al (2018) Neurodevelopmental phenotype caused by a de novo PTPN4 single nucleotide variant disrupting protein localization in neuronal dendritic spines. Clin Genet 94(6):581–585. https://doi.org/10.1111/cge.13450
Bitetto G, Di Fonzo A (2020) Nucleo-cytoplasmic transport defects and protein aggregates in neurodegeneration. Transl Neurodegener 9(1):25. https://doi.org/10.1186/s40035-020-00205-2
Nguyen JM, Qualmann KJ, Okashah R, Reilly A, Alexeyev MF, Campbell DJ (2015) 5p deletions: current knowledge and future directions. Am J Med Genet C Semin Med Genet 169(3):224–238. https://doi.org/10.1002/ajmg.c.31444
Phelan MC (2008) Deletion 22q13.3 syndrome. Orphanet J Rare Dis 3:14. https://doi.org/10.1186/1750-1172-3-14
Phelan K, McDermid HE (2012) The 22q13.3 deletion syndrome (Phelan-McDermid syndrome). Mol Syndromol 2(3–5):186–201. https://doi.org/10.1159/000334260
Bacchelli E et al (2015) Analysis of CHRNA7 rare variants in autism spectrum disorder susceptibility. Am J Med Genet A 167A(4):715–723. https://doi.org/10.1002/ajmg.a.36847
Uhlen M et al (2010) Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28(12):1248–1250. https://doi.org/10.1038/nbt1210-1248
Li H et al (2016) Protein prenylation constitutes an endogenous brake on axonal growth. Cell Rep 16(2):545–558. https://doi.org/10.1016/j.celrep.2016.06.013
Lin Z, Li J, Ji T, Wu Y, Gao K, Jiang Y (2021) ATP1A1 de novo mutation-related disorders: clinical and genetic features. Front Pediatr 9:657256. https://doi.org/10.3389/fped.2021.657256
Guillen Sacoto MJ et al (2020) De novo variants in the ATPase module of MORC2 cause a neurodevelopmental disorder with growth retardation and variable craniofacial dysmorphism. Am J Hum Genet 107(2):352–363. https://doi.org/10.1016/j.ajhg.2020.06.013
Acknowledgements
The authors are thankful to all patients and their families for participating in the study. We thank Emile Myron D Verano for the assistance in data compilation.
Funding
This research was funded by including College of Medicine at Mohammed Bin Rashid University of Medicine and Health Sciences, grant number MBRU-CM-RG2018-04 and MBRU-CM-RG2020-12, Sandooq Al Watan grant (SWARD-F2018-002), and Al Jalila Foundation. Dr. Nasna Nassir was supported by the MBRU Post-Doctoral Fellow Award (MBRU-PD-2020–02).
Author information
Authors and Affiliations
Contributions
Conceptualization: N.N., M.U., and A.A.B.; methodology: N.N., I.S., S.A.S., and A.A.B.; software: N.N., H.A., and A.A; formal analysis: N.N., M.U., A.A.B., I.S., S.A.S., O.A., H.A., M.W.S., A.A.T.; data curation: N.N., I.S., O.A.; writing—original draft preparation: N.N., M.U., A.A.B., M.W.S., I.S.; writing—review and editing: N.N., M.U., A.A.B., M.W.S., A.A.T.
Corresponding authors
Ethics declarations
Ethics approval
This study (AJCH-37) was approved by Dubai Health Authority (DHCA).
Consent to participate
No consent was sought from participants given that the study involved only a de-identified retrospective review of patient data.
Consent for publication
NA
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nassir, N., Sati, I., Al Shaibani, S. et al. Detection of copy number variants and genes by chromosomal microarray in an Emirati neurodevelopmental disorders cohort. Neurogenetics 23, 137–149 (2022). https://doi.org/10.1007/s10048-022-00689-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-022-00689-2