Abstract
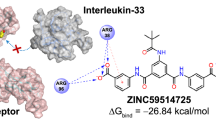
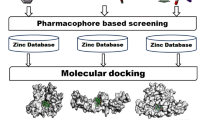
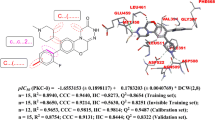
Receptor-interacting protein kinase 2 (RIPK2) plays an essential role in autoimmune response and is suggested as a target for inflammatory diseases. A pharmacophore model was built from a dataset with ponatinib (template) and 18 RIPK2 inhibitors selected from BindingDB database. The pharmacophore model validation was performed by multiple linear regression (MLR). The statistical quality of the model was evaluated by the correlation coefficient (R), squared correlation coefficient (R2), explanatory variance (adjusted R2), standard error of estimate (SEE), and variance ratio (F). The best pharmacophore model has one aromatic group (LEU24 residue interaction) and two hydrogen bonding acceptor groups (MET98 and TYR97 residues interaction), having a score of 24.739 with 14 aligned inhibitors, which were used in virtual screening via ZincPharmer server and the ZINC database (selected in function of the RMSD value). We determined theoretical values of biological activity (logRA) by MLR, pharmacokinetic and toxicology properties, and made molecular docking studies comparing binding affinity (kcal/mol) results with the most active compound of the study (ponatinib) and WEHI-345. Nine compounds from the ZINC database show satisfactory results, yielding among those selected, the compound ZINC01540228, as the most promising RIPK2 inhibitor. After binding free energy calculations, the following molecular dynamics simulations showed that the receptor protein’s backbone remained stable after the introduction of ligands.








Similar content being viewed by others
References
Ferreira LRF, Pestana PR, de Oliveira J, Mesquita-Ferrari RA (2008) Effects of aquatic rehabilitation on symptoms and quality of life in rheumatoid arthritis female patients. Fisioterapia e Pesquisa 15:136–141. https://doi.org/10.1590/S1809-29502008000200005
Mota LMH, Cruz BA, De Albuquerque CP, Gonçalves DP, Laurindo IMM, Pereira IA, De Carvalho JF, Pinheiro GRC, Bertolo MB, Pinto MRC, Louzada-Junior P, Xavier RM, Giorgi RDN, Lima RAC (2015) Update on the 2012 Brazilian Society of Rheumatology Guidelines for the treatment of rheumatoid arthritis: position on the use of tofacitinib. Rev. Bras. Reumatol. 55:512–521. https://doi.org/10.1016/j.rbr.2015.08.004
Chorus AMJ, Miedema HS, Boonen A, Linden SVD (2003) Quality of life and work in patients with rheumatoid arthritis and ankylosing spondylitis of working age. Ann. Rheum. Dis. 62:1178–1184. https://doi.org/10.1136/ard.2002.004861
Tijhuis GJ, Jong Z, Zwinderman AH, Zuijderduin WM, Jansen LMA, Hazes JMW, Vliet Vlieland TPM (2001) The validity of the rheumatoid arthritis quality of life (RAQoL) questionnaire. Rheumatology 40:1112–1119. https://doi.org/10.1093/rheumatology/40.10.1112
Panus PC, Katzung B, Jobst EE, Tinsley SL, Marsters SB, Trevor AJ (2011) Pharmacology for the physical therapist. Artmed, Porto Alegre
Canning P, Ruan Q, Schwerd T, Hrdinka M, Maki JL, Saleh D, Suebsuwong C, Ray S, Brennan PE, Cuny GD, Uhlig HH, Gyrd-Hansen M, Degterev A, Bullock AN (2015) Inflammatory signaling by NOD-RIPK2 is inhibited by clinically relevant type II kinase inhibitors. Chem. Biol. 22:1174–1184. https://doi.org/10.1016/j.chembiol.2015.07.017
Berman J, Westbrook Z, Feng G, Gilliland TN, Bhat H, Weissig IN, Shindyalov Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res. 28:235–242. https://doi.org/10.1093/nar/28.1.235
Gilson MK, Tiqing Liu MB, Nicola G, Hwang L, Chong J (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44:1045–1053. https://doi.org/10.1093/nar/gkv1072
Yamashita S, Furubayashi T, Kataoka M, Sakane T, Sezaki H, Tokuda H (2000) Optimized conditions for prediction of intestinal drug permeability using Caco-2 cells. Eur J Pharmacol 195–204. https://doi.org/10.1016/S0928-0987(00)00076-2
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 16:2785–2791. https://doi.org/10.1002/jcc.21256
Nachbur U, Stafford CA, Bankovacki A, Zhan Y, Lindqvist LM, Fiil BK, Khakham Y, Ko H, Sandow JJ, Falk H, Holien JK, Chau D, Hildebrand J, Vince JE, Sharp PP, Webb AI, Jackman KA, Hlen SM, Kennedy CL, Lowes KN, Murphy JM, Gyrd-Hansen M, Parker MW, Hartland EL, Lew AM, Huang DCS, Lessene G, Silke J (2015) A RIPK2 inhibitor delays NOD signalling events yet prevents inflammatory cytokine production. Nat. Commun. 6:6442. https://doi.org/10.1038/ncomms7442
Santos CBR, Lobato CC, Braga FS, Costa JS, Favacho HAS, Carvalho JCT, Macêdo WJC, Brasil DSB, Silva CHTP, Hage-Melim LIS (2015) Rational design of antimalarial drugs using molecular modeling and statistical analysis. Curr. Pharm. Des. 21:4112–4127. https://doi.org/10.2174/1381612821666150528121423
Santos CBR, Vieira JB, Lobato CC, Hage-Melim LIS, Souto RNP, Lima CS, Costa EVM, Brasil DSB, Macêdo WJC, Carvalho JCT (2013) A SAR and QSAR study of new artemisinin compounds with antimalarial activity. Molecules 19:367–399. https://doi.org/10.3390/molecules19010367
Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson JH (2008) Deterministic pharmacophore detection via multiple flexible alignments of drug-like molecules. J. Comput. Biol. 15:737–754. https://doi.org/10.1089/cmb.2007.0130
Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ (2008) PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 36:223–228. https://doi.org/10.1093/nar/gkn187
Koes DR, Camacho CJ (2012) ZINCPharmer: pharmacophore search of the ZINC database. Nucleic Acids Res. 40:409–414. https://doi.org/10.1093/nar/gks378
Barbosa JP, Ferreira JEV, Figueiredo AF, Almeida RCO, Silva OPP, Carvalho JRC, Cristino MGG, Ciriaco-Pinheiro J, Vieira JLF, Serra RTA (2011) Molecular modeling and chemometric study of anticancer derivatives of artemisinin. J. Serb. Chem. Soc. 76:1263–1282. https://doi.org/10.2298/JSC111227111B
STATISTICA (Data Analysis Software System) (2004) Version 6.1, StatSoft, Inc.
Cunha EL, Santos CF, Braga FS, Costa JS, Silva RC, Favacho HAS, Hage-Melim LIS, Carvalho JCT, Silva CHTP, Santos CBR (2015) Computational investigation of antifungal compounds using molecular modeling and prediction of ADME/Tox properties. J. Comput. Theor. Nanosci. 12:3682–3691. https://doi.org/10.1166/jctn.2015.4260
Harris PA, Bandyopadhyay D, Berger SB, Campobasso N, Capriotti CA, Cox JA, Dare L, Finger JN, Hoffman SJ, Kahler KM, Lehr R, Lich JD, Nagilla R, Nolte RT, Ouellette MT, Pao CS, Schaeffer MC, Smallwood A, Sun HH, Swift BA, Totoritis RD, Ward P, Marquis RW, Bertin J, Gough PJ (2013) Discovery of small molecule RIP1 kinase inhibitors for the treatment of pathologies associated with necroptosis. Med Chem Lett. 4:1238–1243. https://doi.org/10.1021/ml400382p
Xie T, Peng W, Yan C, Wu J, Gong X, Shi Y (2013) Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 5:70–78. https://doi.org/10.1016/j.celrep.2013.08.044
Dassault Systèmes BIOVIA (2015) Discovery studio modeling environment. San Diego
Dassault Systèmes BIOVIA (2018) Discovery Studio 4.0, San Diego
Jayaram B, Sprous D, Beveridge DL (1998) Solvation free energy of biomacromolecules: parameters for a modified generalized born model consistent with the AMBER force field. J. Phys. Chem. B 102:9571–9576. https://doi.org/10.1021/jp982007x
Massova I, Kollman PA (2000) Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discovery Des. 18:113–135. https://doi.org/10.1023/A:1008763014207
Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA (1998) Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helixes. J. Am. Chem. Soc. 120:9401–9409. https://doi.org/10.1021/ja981844
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Scott WRP, Hünenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Kruger P, Van Gunsteren WF (1999) The GROMOS biomolecular simulation program package. J. Phys. Chem. A 103:3596–3607. https://doi.org/10.1021/jp984217f
Mark P, Nilsson L (2001) Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 105:9954–9960. https://doi.org/10.1021/jp003020w
Van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 10:255–262. https://doi.org/10.1007/BF00355047
Lemkul JA, Allen WJ, Bevan DR (2010) Practical considerations for building GROMOS-compatible small-molecule topologies. J. Chem. Inf. Model. 50:2221–2235. https://doi.org/10.1021/ci100335w
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Pereira ALE, Santos GBF, Franco MSF, Federico LB, Silva CHTP, Santos CBR (2017) Molecular modeling and statistical analysis in the design of derivatives of human dipeptidyl peptidase IV. J. Biomol. Struct. Dyn. 36:318–334. https://doi.org/10.1080/07391102.2016.1277163
Birck MG, Campos LJ, Melo EB (2016) Estudo computacional de 1h-imidazol-2-il-pirimidina-4,6-diaminas para a identificação de potenciais precursores de novos agentes antimaláricos. Quim Nova 39:567–574. https://doi.org/10.5935/0100-4042.20160065
Gupta S, Mohan CG (2014) Dual binding site and selective acetylcholinesterase inhibitors derived from integrated pharmacophore models and sequential virtual screening. Biomed. Res. Int. 2014:1–21. https://doi.org/10.1155/2014/291214
Vieira JB, Braga FS, Lobato CC (2014) A QSAR, pharmacokinetic and toxicological study of new artemisinin compounds with anticancer activity. Molecules 19:670–679. https://doi.org/10.3390/molecules190810670
Ames BN, Gurney EG, Miller AJ, Bartsch H (1972) Carcinogens as frameshift mutagens: metabolites and derivatives of 2-acetylaminofluorene and other aromatic amine carcinogens. Proc. Natl. Acad. Sci. U. S. A. 69:3128–3132. https://doi.org/10.1073/pnas.69.11.3128
Silva NSR, Santos CF, Gonçalves LKS, Braga FS, Almeida JR, Lima CS, Brasil DSB, Silva CHTP, Hage-Melim LIS, Santos CBR (2015) Molecular modeling of the major compounds of Sesquiterpenes class in copaiba oil-resin. Br. J. Pharm. Res. 7:247–263. https://doi.org/10.9734/BJPR/2015/17591
Cruz JV, Neto MFA, Silva LB, Ramos RS, Costa JS, Brasil DSB, Lobato CC, Costa GV, Bittencourt JAHM, Silva CHTP, Leite FHA, Santos CBR (2018) Identification of novel protein kinase receptor type 2 inhibitors using pharmacophore and structure-based virtual screening. Molecules 23:453. https://doi.org/10.3390/molecules23020453
Dos Santos CBR, da Silva RR, Ortiz BLS, da Silva GM, Giuliatti S, Balderas-Lopez JL, Navarrete A, Carvalho JCT (2018) Oil from the fruits of Pterodon emarginatus Vog.: a traditional anti-inflammatory. Study combining in vivo and in silico. J. Ethnopharmacol. 222:107–120. https://doi.org/10.1016/j.jep.2018.04.041
Najjar M, Suebsuwong C, Ray SS, Thapa RJ, Maki JL, Nogusa S, Shah S, Saleh D, Gough PJ, Bertin J, Yuan J, Balachandran S, Cuny GD, Degterev A (2015) Structure guided design of potent and selective ponatinib-based hybrid inhibitors for RIPK1. Cell Rep. 10:1850–1860. https://doi.org/10.1016/j.celrep.2015.02.052
Ou-Yang S-s, Lu J-y, Kong X-q, Liang Z-j, Luo C, Hualiang J (2012) Computational drug discovery. Acta Pharmacol. Sin. 33:1131–1140. https://doi.org/10.1038/aps.2012.109
Acknowledgements
We would like to acknowledge the computing time provided on the Blue Gene/Q supercomputer supported by the Center for Research Computing (Rice University), Superintendência de Tecnologia da Informação da Universidade de São Paulo, Programa de Pós-graduação em Ciências Farmacêuticas of Universidade Federal do Amapá, Brazil and to the Laboratório de Modelagem e Química Computacional (LMQC/UNIFAP).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare no financial conflicts of interest.
Rights and permissions
About this article
Cite this article
Cruz, J.V., Serafim, R.B., da Silva, G.M. et al. Computational design of new protein kinase 2 inhibitors for the treatment of inflammatory diseases using QSAR, pharmacophore-structure-based virtual screening, and molecular dynamics. J Mol Model 24, 225 (2018). https://doi.org/10.1007/s00894-018-3756-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3756-y