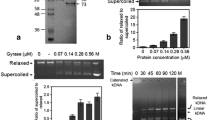
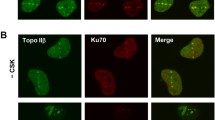
Abstract
Topoisomerases are crucial enzymes in genome maintenance that modulate the topological changes during DNA metabolism. Deinococcus radiodurans, a Gram-positive bacterium is characterized by its resistance to many abiotic stresses including gamma radiation. Its multipartite genome encodes both type I and type II topoisomerases. Time-lapse studies using fluorescently tagged topoisomerase IB (drTopoIB-RFP) and DNA gyrase (GyrA-RFP) were performed to check the dynamics and localization with respect to DNA repair and cell division under normal and post-irradiation growth conditions. Results suggested that TopoIB and DNA gyrase are mostly found on nucleoid, highly dynamic, and show growth phase-dependent subcellular localization. The drTopoIB-RFP was also present at peripheral and septum regions but does not co-localize with the cell division protein, drFtsZ. On the other hand, DNA gyrase co-localizes with PprA a pleiotropic protein involved in radioresistance, on the nucleoid during the post-irradiation recovery (PIR). The topoIB mutant was found to be sensitive to hydroxyurea treatment, and showed more accumulation of single-stranded DNA during the PIR, compared to the wild type suggesting its role in DNA replication stress. Together, these results suggest differential localization of drTopoIB-RFP and GyrA-RFP in D. radiodurans and their interaction with PprA protein, emphasizing the functional significance and role in radioresistance.






Similar content being viewed by others
References
Ahmed W, Sala C, Hegde SR, Jha RK, Cole ST, Nagaraja V (2017) Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet 13(5):e1006754. https://doi.org/10.1371/journal.pgen.1006754. (PMID:28463980;PMCID:PMC5433769)
Aubry A, Fisher LM, Jarlier V, Cambau E (2006) First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem Biophys Res Commun 348(1):158–165. https://doi.org/10.1016/j.bbrc.2006.07.017. (Epub 2006 Jul 13 PMID: 16876125)
Bouige A, Darmon A, Piton J, Roue M, Petrella S, Capton E, Forterre P, Aubry A, Mayer C (2013) Mycobacterium tuberculosis DNA gyrase possesses two functional GyrA-boxes. Biochem J 455(3):285–294. https://doi.org/10.1042/BJ20130430. (PMID: 23869946)
Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70:369–413
Chen SH, Chan NL, Hsieh TS (2013) New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170
Cox MM, Battista JR (2005) Deinococcus radiodurans—the consummate survivor. Nat Rev Microbiol 3(11):882–892
de la Tour CB, Passot FM, Toueille M, Mirabella B, Guérin P, Blanchard L, Servant P, de Groot A, Sommer S, Armengaud J (2013) Comparative proteomics reveals key proteins recruited at the nucleoid of Deinococcus after irradiation-induced DNA damage. Proteomics 23–24:3457–3469. https://doi.org/10.1002/pmic.201300249. (Epub 2013 Dec 4 PMID: 24307635)
Devigne A, Mersaoui S, Bouthier-de-la-Tour C, Sommer S, Servant P (2013) The PprA protein is required for accurate cell division of γ-irradiated Deinococcus radiodurans bacteria. DNA Repair (Amst). 12(4):265–72. https://doi.org/10.1016/j.dnarep.2013.01.004
Devigne A, Guérin P, Lisboa J, Quevillon-Cheruel S, Armengaud J, Sommer S, Bouthier de la Tour C, Servant P (2016) PprA protein is involved in chromosome segregation via its physical and functional interaction with DNA gyrase in irradiated deinococcus radiodurans bacteria. mSphere 1(1):e00036-15. https://doi.org/10.1128/mSphere.00036-15. (PMID: 27303692; PMCID: PMC4863600)
Floc’h K, Lacroix F, Servant P, Wong YS, Kleman JP, Bourgeois D, Timmins J (2019) Cell morphology and nucleoid dynamics in dividing deinococcus radiodurans. Nat Commun 10(1):3815. https://doi.org/10.1038/s41467-019-11725-5.PMID:31444361;PMCID:PMC6707255
Galli E, Midonet C, Paly E, Barre FX (2017) Fast growth conditions uncouple the final stages of chromosome segregation and cell division in escherichia coli. PLoS Genet 13(3):e1006702. https://doi.org/10.1371/journal.pgen.1006702.PMID:28358835;PMCID:PMC5391129
Kampranis SC, Bates AD, Maxwell A (1999) A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci U S A 96(15):8414–8419. https://doi.org/10.1073/pnas.96.15.8414.PMID:10411889;PMCID:PMC17530
Kota S, Misra HS (2008) Identification of a DNA processing complex from Deinococcus radiodurans. Biochem Cell Biol. 86(5):448–58. https://doi.org/10.1139/o08-122
Kota S, Misra HS (2015) Topoisomerase IB of Deinococcus radiodurans resolves guanine quadruplex DNA structures in vitro. J Biosci. 40:833–843. https://doi.org/10.1007/s12038-015-9571-z
Kota S, Charaka VK, Misra HS (2014a) PprA, a pleiotropic protein for radioresistance, works through DNA gyrase and shows cellular dynamics during postirradiation recovery in Deinococcus radiodurans. J Genet 93(2):349–354. https://doi.org/10.1007/s12041-014-0382-z. (PMID: 25189229)
Kota S, Charaka VK, Misra HS (2014) PprA, a pleiotropic protein for radioresistance, works through DNA gyrase and shows cellular dynamics during postirradiation recovery in Deinococcus radiodurans. J Genet. 93(2):349–54
Kota S, Rajpurohit YS, Charaka V, Satoh K, Narumi I, Misra HS (2016) DNA gyrase of deinococcus radiodurans is characterized as type II bacterial topoisomerase and its activity is differentially regulated by PprA in vitro. Extremophile 20:195–205
Kota S, Chaudhary R, Mishra S, Misra HS (2021) Topoisomerase IB interacts with genome segregation proteins and is involved in multipartite genome maintenance in Deinococcus radiodurans. Microbiol Res 242:126609
Kramlinger VM, Hiasa H (2006) The, “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J Biol Chem 281(6):3738–3742. https://doi.org/10.1074/jbc.M511160200. (Epub 2005 Dec 5 PMID: 16332690)
Krogh BO, Shuman S (2002) A poxvirus-like type IB topoisomerase family in bacteria. Proc Natl Acad Sci USA. 99(4):1853–8. https://doi.org/10.1073/pnas.032613199. (Epub 2002)
Levin-Zaidman S, Englander J, Shimoni E, Sharma AK, Minton KW (2003) A: Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299(5604):254–256. https://doi.org/10.1126/science.1077865
Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, Wu L, Thompson DK, Xu D, Rogozin IB, Gaidamakova EK, Zhai M, Makarova KS, Koonin EV, Daly MJ (2003) Transcriptome dynamics of deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci USA. 100(7):4191–6. https://doi.org/10.1073/pnas.0630387100
Maier RM (2009) Bacterial growth. In: Maier RM, Pepper IL, Gerb CP (eds) Environmental toxicology, part I: review of basic microbiological concepts, 2nd edn. Elsevier Academic Press, San Diego, CA, pp 37–54
Maurya GK, Kota S, Misra HS (2019) Characterisation of ParB encoded on multipartite genome in Deinococcus radiodurans and their roles in radioresistance. Microbiol Res 223:22–32
Maurya GK, Chaudhary R, Pandey N, Misra HS (2021) Molecular insights into replication initiation in a multipartite genome harboring bacterium Deinococcus radiodurans. J Biol Chem 296:100451. https://doi.org/10.1016/j.jbc.2021.100451. (Epub 2021 Feb 21)
Mishra S, Misra HS, Kota S (2022) FtsK, a DNA Motor protein, coordinates the genome segregation and early cell division processes in deinococcus radiodurans. mBio. 13(6):e0174222. https://doi.org/10.1128/mbio.01742-22
Misra HS, Khairnar NP, Kota S, Shrivastava S, Joshi VP, Apte SK (2006) An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol Microbiol 59(4):1308–1316
Misra HS, Rajpurohit YS, Kota S (2013) Physiological and molecular basis of extreme radioresistance in Deinococcus radiodurans. Curr Sci 104:194–206
Modi KM, Tewari R, Misra HS (2014) FtsZDr, a tubulin homologue in radioresistant bacterium deinococcus radiodurans is characterized as a GTPase exhibiting polymerization/depolymerization dynamics in vitro and FtsZ ring formation in vivo. Int J Biochem Cell Biol 50:38–46. https://doi.org/10.1016/j.biocel.2014.01.015
Patel A, Shuman S, Mondragón A (2006) Crystal structure of a bacterial type IB DNA topoisomerase reveals a preassembled active site in the absence of DNA. J Biol Chem 281(9):6030–6037. https://doi.org/10.1074/jbc.M512332200. (Epub 2005 Dec 19 PMID: 16368685)
Patel A, Yakovleva L, Shuman S, Mondragón A (2010) Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site. Structure 18(6):725–733. https://doi.org/10.1016/j.str.2010.03.007.PMID:20541510;PMCID:PMC2886027
Pommier Y, Nussenzweig A, Takeda S et al (2022) Human topoisomerases and their roles in genome stability and organization. Nat Rev Mol Cell Biol 23:407–427
Ptacin JL, Shapiro L (2013) Chromosome architecture is a key element of bacterial cellular organization. Cell Microbiol. 1:45–52. https://doi.org/10.1111/cmi.12049. (Epub 2012, PMID: 23078580; PMCID: PMC3660146)
Schäfer M, Schmitz C, Facius R, Horneck G, Milow B, Funken KH, Ornery J (2000) Systematic study of parameters influencing the action of Rose Bengal with visible light on bacterial cells: comparison between the biological effect and singletoxygen production. Photochem Photobiol 71(2000):514–523
Slade D, Radman M (2011) Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 75:133–191
Slade D, Lindner AB, Paul G, Radman M (2009) Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136(6):1044–1055. https://doi.org/10.1016/j.cell.2009.01.018. (PMID: 19303848)
Soren BC, Dasari JB, Ottaviani A, Iacovelli F, Fiorani P (2020) Topoisomerase IB: a relaxing enzyme for stressed DNA. Cancer Drug Resist 3(1):18–25. https://doi.org/10.20517/cdr.2019.106.PMID:35582040;PMCID:PMC9094055
Stracy M, Wollman AJM, Kaja E, Gapinski J, Lee J-E, Leek VA, McKie SJ, Mitchenall LA, Maxwell A, Sherratt DJ, Leake MC, Zawadzki P (2019) Single-molecule imaging of DNA gyrase activity in living Escherichia coli. Nucleic Acids Res 47(1):210–220. https://doi.org/10.1093/nar/gky1143
Tadesse S, Graumann PL (2006) Differential and dynamic localization of topoisomerases in Bacillus subtilis. J Bacteriol 188(8):3002–3011. https://doi.org/10.1128/JB.188.8.3002-3011.2006.PMID:16585761;PMCID:PMC1446999
Takahashi DT, Gadelle D, Agama K, Kiselev E, Zhang H, Yab E, Petrella S, Forterre P, Pommier Y, Mayer C (2022) Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states. Nat Commun 13(1):59. https://doi.org/10.1038/s41467-021-27686-7.PMID:35013228;PMCID:PMC8748870
White, O., Eisen, J.A., Heidelberg, J.F., Hickey, E.K., Peterson, J.D., Dodson, R.J., et al. (1999). Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 2861571–1577.
Wuqiang Huang, Zhiping Liu, Yikang S Rong. (2021) Dynamic localization of DNA topoisomerase I and its functional relevance during Drosophila development. G3 Genes Genomes Genetics https://doi.org/10.1093/g3journal/jkab202
Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443(7111):569–573. https://doi.org/10.1038/nature05160. (Epub 2006 Sep 27 PMID: 17006450)
Zawadzki P, Stracy M, Ginda K, Zawadzka K, Lesterlin C, Kapanidis AN, Sherratt DJ (2015) The localization and action of topoisomerase IV in escherichia coli chromosome segregation is coordinated by the SMC complex. MukBEF. Cell Rep. 13(11):2587–2596. https://doi.org/10.1016/j.celrep.2015.11.034
Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR (2000) Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem 275(11):8103–8113. https://doi.org/10.1074/jbc.275.11.8103. (PMID: 10713132)
Acknowledgements
We sincerely thank Dr. Hema Rajaram, Bhabha Atomic Research Centre, Mumbai, for her suggestions and support while preparing the manuscript. Miss Himani Tewari is grateful to the Department of Atomic Energy, Government of India, for the research fellowship.
Author information
Authors and Affiliations
Contributions
SM performed the experiments, analyzed the results, and prepared the manuscript. HT performed the experiments and manuscript preparation. RC conducted the experiments and manuscript preparation. HSM analyzed the data and manuscript preparation. SK conceived the idea, planned and conducted the experiments, analyzed the data, wrote the manuscript, and communicated for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Communicated by Atomi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
792_2023_1323_MOESM1_ESM.pdf
Supplementary file1 Fig S1: Growth studies of D. radiodurans cells expressing RFP fusions of TopoIB and DNAgyrA episomally. Different dilutions of exponentially growing cells of D. radiodurans (R1) expressing TopoIB-RFP and DNAgyrA-RFP under unirradiated (UIrr) and post-irradiation (Irr) were spotted on TYG agar plates. Fig S2: Time-lapse microscopy of a population of D. radiodurans cells expressing TopoIB-RFP. Microscopic images were taken in differential inference contrast (DIC) and TRITC (TopoIB-RFP- red) channels. The scale bar is 1 μm. Fig S3: Time-lapse microscopy of a population of D. radiodurans cells expressing DNAgyrA-RFP. Microscopic images were taken in differential inference contrast (DIC) and TRITC (DNAgyrA-RFP-red) channels. The scale bar is 1 μm. Fig S4: Growth studies of D. radiodurans (R1) and topo:nptII mutant (Δtopo) under unirradiated (UIrr), post-irradiated (Irr) conditions, post-hydroxyurea (HU) treatment (HU-UIrr) and post-HU treatment followed by irradiation (HU-Irr). Different dilutions of exponentially growing cells were spotted on TYG agar plates. Fig S5: Microscopic images of D. radiodurans cells expressing drDNAgyrA-RFP after hydroxyurea (HU) treatment. Cells were grown in TYG broth and monitored under the microscope at specific time intervals (t=hrs) post-HU treatment. Fig S6: DNA binding motif analysis of D. radiodurans DNA Gyrase A. Sequence alignment of D. radiodurans DNA Gyrase A C- terminal domain (CTD) with M. tuberculosis DNA Gyrase A CTD was done using PROMALS3D (a). A schematic representation of the conserved DNA binding motif GyrA-box and a unique GyrA-box-I at the GyrA-CTD is given (b). Boundaries of the motifs are marked as a black box for GyrA-box and a red box for GyrA-box-I. (PDF 1088 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, S., Tewari, H., Chaudhary, R. et al. Differential cellular localization of DNA gyrase and topoisomerase IB in response to DNA damage in Deinococcus radiodurans. Extremophiles 28, 7 (2024). https://doi.org/10.1007/s00792-023-01323-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00792-023-01323-1