Abstract
Objectives
The aim of this systematic review and meta-analysis is to assess the diagnostic potential of salivary metabolomics in the detection of oral potentially malignant disorders (OPMDs) and oral cancer (OC).
Materials and methods
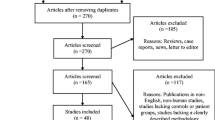
A systematic review was performed in accordance with the 3rd edition of the Centre for Reviews and Dissemination (CRD) and Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement. Electronic searches for articles were carried out in the PubMed, Web of Science, and Scopus databases. The quality assessment of the included studies was evaluated using the Newcastle-Ottawa Quality Assessment Scale (NOS) and the new version of the QUADOMICS tool. Meta-analysis was conducted whenever possible. The effect size was presented using the Forest plot, whereas the presence of publication bias was examined through Begg’s funnel plot.
Results
A total of nine studies were included in the systematic review. The metabolite profiling was heterogeneous across all the studies. The expression of several salivary metabolites was found to be significantly altered in OPMDs and OCs as compared to healthy controls. Meta-analysis was able to be conducted only for N-acetylglucosamine. There was no significant difference (SMD = 0.15; 95% CI − 0.25–0.56) in the level of N-acetylglucosamine between OPMDs, OC, and the control group.
Conclusion
Evidence for N-acetylglucosamine as a salivary biomarker for oral cancer is lacking. Although several salivary metabolites show changes between healthy, OPMDs, and OC, their diagnostic potential cannot be assessed in this review due to a lack of data. Therefore, further high-quality studies with detailed analysis and reporting are required to establish the diagnostic potential of the salivary metabolites in OPMDs and OC.
Clinical relevance
While some salivary metabolites exhibit significant changes in oral potentially malignant disorders (OPMDs) and oral cancer (OC) compared to healthy controls, the current evidence, especially for N-acetylglucosamine, is inadequate to confirm their reliability as diagnostic biomarkers. Additional high-quality studies are needed for a more conclusive assessment of salivary metabolites in oral disease diagnosis.


Similar content being viewed by others
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Sankaranarayanan R, Ramadas K, Amarasinghe H, Subramanian S, Johnson N (2015) In: Gelband H, Jha P, Sankaranarayanan R, Horton S (eds) Cancer: Disease Control Priorities, Third Edition (Volume 3). The International Bank for Reconstruction and Development / The World Bank© 2015 International Bank for Reconstruction and Development / The World Bank, Washington (DC)
Farah CS, Woo S-B, Zain RB, Sklavounou A, McCullough MJ, Lingen M (2014) Oral cancer and oral potentially malignant disorders. Int J Dent 2014:853479
Warnakulasuriya S, Kujan O, Aguirre-Urizar JM, Bagan JV, González-Moles M, Kerr AR et al (2021) Oral potentially malignant disorders: a consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis 27(8):1862–1880
Warnakulasuriya S, Kujan O, Aguirre-Urizar JM, Bagan JV, González-Moles MÁ, Kerr AR et al (2021) Oral potentially malignant disorders: a consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis 27(8):1862–1880
Pandarathodiyil AK, Vijayan SP, Milanes D, Chopra V, Anil S (2022) Adjunctive techniques and diagnostic aids in the early detection of oral premalignant disorders and cancer: an update for the general dental practitioners. J Pharm Bioall Sci 14(5):28
Jayasinghe RD, Hettiarachchi P, Amugoda D, Kumaraarachchi M, Liyanage R, Siriwardena B et al (2020) Validity of Toluidine blue test as a diagnostic tool for high risk oral potentially malignant disorders- a multicentre study in Sri Lanka. J Oral Biol Craniofac Res 10(4):547–551
Rashid A, Warnakulasuriya S (2015) The use of light-based (optical) detection systems as adjuncts in the detection of oral cancer and oral potentially malignant disorders: a systematic review. J Oral Pathol Med 44(5):307–328
Dai X, Shen L (2022) Advances and trends in omics technology development. Front Med (Lausanne) 9:911861
Kosmides AK, Kamisoglu K, Calvano SE, Corbett SA, Androulakis IP (2013) Metabolomic fingerprinting: challenges and opportunities. Crit Rev Biomed Eng 41(3):205–221
Kessler AT, Bhatt AA (2018) Review of the major and minor salivary glands, part 1: anatomy, infectious, and inflammatory processes. J Clin Imaging Sci 8:47
Ghannam MG, Singh P (2022) StatPearls. StatPearls PublishingCopyright © 2022, StatPearls Publishing LLC, Treasure Island (FL)
Nijakowski K, Gruszczyński D, Kopała D, Surdacka A (2022) Salivary metabolomics for oral squamous cell carcinoma diagnosis: a systematic review. Metabolites 12(4):294
Patil DJ, More CB (2021) Salivary metabolomics – a diagnostic and biologic signature for oral cancer. J Oral Maxillofac Surg Med Pathol 33(5):546–554
Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, Suksuwan W et al (2021) Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect 27(2):285.e1-.e4
Huan T, Tran T, Zheng J, Sapkota S, MacDonald SW, Camicioli R et al (2018) Metabolomics analyses of saliva detect novel biomarkers of Alzheimer’s disease. J Alzheimers Dis 65(4):1401–1416
Sakanaka A, Kuboniwa M, Katakami N, Furuno M, Nishizawa H, Omori K et al (2021) Saliva and plasma reflect metabolism altered by diabetes and periodontitis. Front Mol Biosci 8:742002
Shao Y, Li T, Liu Z, Wang X, Xu X, Li S et al (2021) Comprehensive metabolic profiling of Parkinson’s disease by liquid chromatography-mass spectrometry. Mol Neurodegener 16(1):4
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD et al (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372:n71
Ishikawa S, Sugimoto M, Edamatsu K, Sugano A, Kitabatake K, Iino M (2020) Discrimination of oral squamous cell carcinoma from oral lichen planus by salivary metabolomics. Oral Dis 26(1):35–42
Rekha P, Aruna P, Brindha E, Koteeswaran D, Baludavid M, Ganesan S (2016) Near-infrared Raman spectroscopic characterization of salivary metabolites in the discrimination of normal from oral premalignant and malignant conditions. J Raman Spectrosc 47(7):763–772
Shigeyama H, Wang T, Ichinose M, Ansai T, Lee SW (2019) Identification of volatile metabolites in human saliva from patients with oral squamous cell carcinoma via zeolite-based thin-film microextraction coupled with GC-MS. J Chromatogr B Analyt Technol Biomed Life Sci 1104:49–58
Song X, Yang X, Narayanan R, Shankar V, Ethiraj S, Wang X et al (2020) Oral squamous cell carcinoma diagnosed from saliva metabolic profiling. Proc Natl Acad Sci U S A 117(28):16167–16173
Sridharan G, Ramani P, Patankar S, Vijayaraghavan R (2019) Evaluation of salivary metabolomics in oral leukoplakia and oral squamous cell carcinoma. J Oral Pathol Med 48(4):299–306
Wei JE, Xie GX, Zhou ZT, Shi P, Qiu YP, Zheng XJ et al (2011) Salivary metabolite signatures of oral cancer and leukoplakia. Int J Cancer 129(9):2207–2217
Yan SK, Wei BJ, Lin ZY, Yang Y, Zhou ZT, Zhang WD (2008) A metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral Oncol 44(5):477–483
Kitabatake K, Ishikawa S, Sugimoto M, Enomoto A, Kaneko M, Ota S et al (2023) Salivary metabolomics for oral leukoplakia with and without dysplasia. J Stomatol Oral Maxillofac Surg 124(6s):101618
Ishikawa S, Wong DTW, Sugimoto M, Gleber-Netto FO, Li F, Tu M et al (2019) Identification of salivary metabolites for oral squamous cell carcinoma and oral epithelial dysplasia screening from persistent suspicious oral mucosal lesions. Clin Oral Invest 23(9):3557–3563
Didangelos A, Yin X, Mandal K, Baumert M, Jahangiri M, Mayr M (2010) Proteomics characterization of extracellular space components in the human aorta. Mol Cell Proteomics 9(9):2048–2062
McHale CM, Zhang L, Thomas R, Smith MT (2013) Analysis of the transcriptome in molecular epidemiology studies. Environ Mol Mutagen 54(7):500–517
Liberti MV, Locasale JW (2016) The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 41(3):211–218
Zhou D, Duan Z, Li Z, Ge F, Wei R, Kong L (2022) The significance of glycolysis in tumor progression and its relationship with the tumor microenvironment. Front Pharmacol 13:1091779
Zhao H, Hu CY, Chen WM, Huang P (2019) Lactate promotes cancer stem-like property of oral sequamous cell carcinoma. Curr Med Sci 39(3):403–409
Tołoczko-Iwaniuk N, Dziemiańczyk-Pakieła D, Celińska-Janowicz K, Zaręba I, Klupczyńska A, Kokot ZJ et al (2020) Proline-dependent induction of apoptosis in oral squamous cell carcinoma (OSCC)-the effect of celecoxib. Cancers (Basel) 12(1):136
Morgan AA, Rubenstein E (2013) Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS ONE 8(1):e53785
Phang JM, Liu W, Hancock CN, Fischer JW (2015) Proline metabolism and cancer: emerging links to glutamine and collagen. Curr Opin Clin Nutr Metab Care 18(1):71–77
Novita Sari I, Setiawan T, Seock Kim K, Toni Wijaya Y, Won Cho K, Young KH (2021) Metabolism and function of polyamines in cancer progression. Cancer Lett 519:91–104
Manni A, Grove R, Kunselman S, Aldaz M (1995) Involvement of the polyamine pathway in breast cancer progression. Cancer Lett 92(1):49–57
Gilmour SK (2007) Polyamines and nonmelanoma skin cancer. Toxicol Appl Pharmacol 224(3):249–256
Upp JR Jr, Saydjari R, Townsend CM Jr, Singh P, Barranco S, Thompson JC (1988) Polyamine levels and gastrin receptors in colon cancers. Ann Surg 207(6):662
Gupta S, Ahmad N, Marengo SR, MacLennan GT, Greenberg NM, Mukhtar H (2000) Chemoprevention of prostate carcinogenesis by α-difluoromethylornithine in TRAMP mice. Can Res 60(18):5125–5133
Harper A, Miller R, Block K (1984) Branched-chain amino acid metabolism. Annu Rev Nutr 4(1):409–454
Jung MK, Okekunle AP, Lee JE, Sung MK, Lim YJ (2021) Role of branched-chain amino acid metabolism in tumor development and progression. J Cancer Prev 26(4):237–243
Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL et al (2014) O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell 54(5):820–831
Kamigaito T, Okaneya T, Kawakubo M, Shimojo H, Nishizawa O, Nakayama J (2014) Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis 17(1):18–22
Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X et al (2011) O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochimica et Biophysica Acta (BBA) - Mol Basis Dis 1812(4):514–9
Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA III et al (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337(6097):975–980
Zhu Q, Zhou L, Yang Z, Lai M, Xie H, Wu L et al (2012) O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med Oncol 29(2):985–993
Rozanski W, Krzeslak A, Forma E, Brys M, Blewniewski M, Wozniak P et al (2012) Prediction of bladder cancer based on urinary content of MGEA5 and OGT mRNA level. Clin Lab 58(5):579
Ma Z, Vocadlo DJ, Vosseller K (2013) Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J Biol Chem 288(21):15121–15130
Shi Y, Tomic J, Wen F, Shaha S, Bahlo A, Harrison R et al (2010) Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia 24(9):1588–1598
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–74
García-Gómez R, Bustelo XR, Crespo P (2018) Protein–protein interactions: emerging oncotargets in the RAS-ERK pathway. Trends Cancer 4(9):616–633
Sodi VL, Khaku S, Krutilina R, Schwab LP, Vocadlo DJ, Seagroves TN et al (2015) mTOR/MYC axis regulates O-GlcNAc transferase expression and O-GlcNAcylation in breast cancer c-MYC regulates OGT expression in cancer cells. Mol Cancer Res 13(5):923–933
Subramani R, Poudel S, Smith KD, Estrada A, Lakshmanaswamy R (2022) Metabolomics of breast cancer: a review. Metabolites 12(7):643
Kdadra M, Höckner S, Leung H, Kremer W, Schiffer E (2019) Metabolomics biomarkers of prostate cancer: a systematic review. Diagnostics (Basel) 9(1):21
Taylor NJ, Gaynanova I, Eschrich SA, Welsh EA, Garrett TJ, Beecher C et al (2020) Metabolomics of primary cutaneous melanoma and matched adjacent extratumoral microenvironment. PLoS ONE 15(10):e0240849
Bull FC, Al-Ansari SS, Biddle S, Borodulin K, Buman MP, Cardon G et al (2020) World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br J Sports Med 54(24):1451–1462
Seematter G, Binnert C, Tappy L (2005) Stress and metabolism. Metab Syndr Relat Disord 3(1):8–13
Guasch-Ferré M, Bhupathiraju SN, Hu FB (2018) Use of metabolomics in improving assessment of dietary intake. Clin Chem 64(1):82–98
Funding
This study is funded by the Dental Postgraduate Research Grant (DPRG), Faculty of Dentistry, Universiti Malaya [UMG003E-2023 & UMG022E-2022].
Author information
Authors and Affiliations
Contributions
NSMN, AR, WMN made substantial contribution to conception of the study. NSMN and AR contributed to the study selection. NSMN, WMN and AR collected, extracted and managed data. NSMN, FR, KK, NES and MSH performed data processing, interpretation and assessed study quality and risk of bias. PSJ conducted data analysis and strategy for data synthesis. The manuscript was written by NSMN, AR and WMN, tables were prepared by NSMN, FR and AR, figure 1 was prepared by NSMN, AR and KK and figure 2 and 3 was prepared by PSJ. The GRADE approach was attempted by LD. All authors have read, revised critically, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nazar, N.S.B.M., Ramanathan, A., Ghani, W.M.N. et al. Salivary metabolomics in oral potentially malignant disorders and oral cancer patients—a systematic review with meta-analysis. Clin Oral Invest 28, 98 (2024). https://doi.org/10.1007/s00784-023-05481-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00784-023-05481-6