Abstract
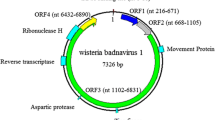
The genome sequence of a novel circular DNA virus related to members of the genus Badnavirus was identified in diseased jujube trees by high-throughput sequencing and verified by conventional Sanger sequencing of cloned PCR products. The name "jujube badnavirus WS" (JuBWS) is proposed for this virus. Diseased jujube leaves showed yellow mosaic and malformation symptoms, with round chlorotic spots found on diseased fruit. The genome of this virus has a length of 6450 nt and has a typical badnavirus genomic structure with three open reading frames (ORFs). JuBWS was identified as a novel badnavirus based on nucleotide differences in the RNase (RT + RNase H) coding region of ORF3. The JuBWS sequence showed 70.48–76.41% nucleotide sequence identity to other known badnaviruses, thus meeting the taxonomic criterion for establishing a new species within the genus Badnavirus. This study suggested that the novel badnavirus might be a pathogen associated with jujube mosaic disease, and this will be investigated in the future.

Similar content being viewed by others
References
Liu MJ, Liu P, Liu GN (2013) Advances of research on germplasm resources of Chinese jujube. Acta Hortic. https://doi.org/10.17660/ActaHortic.2013.993.1
Du K, Liu S, Chen Z et al (2017) Full genome sequence of jujube mosaic-associated virus, a new member of the family Caulimoviridae. Arch Virol 162:3221–3224. https://doi.org/10.1007/s00705-017-3438-6
Medberry SL, Lockhart BEL, Olszewski NE (1990) Properties of Commelina yellow mottle virus’s complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucl Acids Res 18:5505–5513. https://doi.org/10.1093/nar/18.18.5505
Borah BK, Sharma S, Kant R et al (2013) Bacilliform DNA-containing plant viruses in the tropics: commonalities within a genetically diverse group: tropical badnaviruses and tungroviruses. Mol Plant Pathol 14:759–771. https://doi.org/10.1111/mpp.12046
Chabannes M, Gabriel M, Aksa A et al (2021) Badnaviruses and banana genomes: a long association sheds light on Musa phylogeny and origin. Mol Plant Pathol 22:216–230. https://doi.org/10.1111/mpp.13019
Chambers GA, Donovan NJ, Bodaghi S et al (2018) A novel citrus viroid found in Australia, tentatively named citrus viroid VII. Arch Virol 163:215–218. https://doi.org/10.1007/s00705-017-3591-y
Muller E, Sackey S (2005) Molecular variability analysis of five new complete cacao swollen shoot virus genomic sequences. Arch Virol 150:53–66. https://doi.org/10.1007/s00705-004-0394-8
Tahir MN, Bolus S, Grinstead SC et al (2021) A new virus of the family Tombusviridae infecting sugarcane. Arch Virol 166:961–965. https://doi.org/10.1007/s00705-020-04908-9
Kazmi SA, Yang Z, Hong N et al (2015) Characterization by small RNA sequencing of taro bacilliform CH Virus (TaBCHV), a novel badnavirus. PLoS ONE 10:e0134147. https://doi.org/10.1371/journal.pone.0134147
Marais A, Umber M, Filloux D et al (2020) Yam asymptomatic virus 1, a novel virus infecting yams (Dioscorea spp.) with significant prevalence in a germplasm collection. Arch Virol 165:2653–2657. https://doi.org/10.1007/s00705-020-04787-0
Diaz-Lara A, Mosier NJ, Stevens K et al (2020) Evidence of rubus yellow net virus integration into the red raspberry genome. Cytogenet Genome Res 160:329–334. https://doi.org/10.1159/000509845
Laney AG, Hassan M, Tzanetakis IE (2012) An integrated badnavirus is prevalent in fig germplasm. Phytopathology® 102:1182–1189. https://doi.org/10.1094/PHYTO-12-11-0351
Xu D, Mock R, Kinard G, Li R (2011) Molecular analysis of the complete genomic sequences of four isolates of Gooseberry vein banding associated virus. Virus Genes 43:130–137. https://doi.org/10.1007/s11262-011-0614-8
Teycheney P-Y, Geering ADW, Dasgupta I et al (2020) ICTV virus taxonomy profile: caulimoviridae. J Gen Virol 101:1025–1026. https://doi.org/10.1099/jgv.0.001497
Kidanemariam DB, Sukal AC, Abraham AD et al (2018) Identification and molecular characterization of Taro bacilliform virus and Taro bacilliform CH virus from East Africa. Plant Pathol 67:1977–1986. https://doi.org/10.1111/ppa.12921
Bhat A, Hohn T, Selvarajan R (2016) Badnaviruses: the current global scenario. Viruses 8:177. https://doi.org/10.3390/v8060177
Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. https://doi.org/10.1101/gr.074492.107
Peng Y, Leung HCM, Yiu SM, Chin FYL (2012) IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. https://doi.org/10.1093/bioinformatics/bts174
Bateman A, Coggill P, Finn RD (2010) DUFs: families in search of function. Acta Crystallogr F Struct Biol Cryst Commun 66:1148–1152. https://doi.org/10.1107/S1744309110001685
Acknowledgements
Financial support was provided by the Research and Demonstration of Key Technologies for Blockade Prevention and Control of Jujube Virus Diseases in Xinjiang (grant number: 2018E02026) and “Excellent Postdoctoral Professionals and General Funding Staff of Living Subsidies” from the Department of Human Resources and Social Security of Xinjiang Uygur Autonomous Region.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Elvira Fiallo-Olivé.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, B., Zhang, G., Song, D. et al. Complete genome sequence of a novel virus belonging to the genus Badnavirus in jujube (Ziziphus jujuba Mill.) in China. Arch Virol 167, 1885–1888 (2022). https://doi.org/10.1007/s00705-022-05482-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05482-y