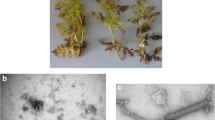
Abstract
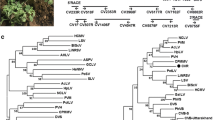
A putative new virus with sequence similarity to members of the genus Cavemovirus in the family Caulimoviridae was identified in wild chicory (Cichorium intybus) by next-generation sequencing (NGS). The putative new virus was tentatively named “chicory mosaic cavemovirus” (ChiMV), and its genome was determined to be 7,775 nucleotides (nt) long with the typical genome organization of cavemoviruses. ORF1 encodes a putative coat protein/movement polyprotein (1,278 aa), ORF2 encodes a putative replicase (650 aa), and ORF3 encodes a putative transactivator factor (384 aa). The first two putative proteins have 46.2% and 68.7% amino acid sequence identity to the CP/MP protein (YP_004347414) and replicase (YP_004347415), respectively, of sweet potato collusive virus (SPCV). ORF3 encodes a protein with 38.5% amino acid sequence identity to the putative transactivator factor (NP_056849) of cassava vein mosaic virus (CsVMV). The new putative viral genome and those of three cavemoviruses (epiphyllum virus 4 [EpV-4], SPCV, and CsVMV) differ by 24-27% in the nt sequence of the replicase gene, which exceeds the species demarcation cutoff (>20%) for the family.


Similar content being viewed by others
References
Van Der Maesen LJG (1996) Asteraceae, cladistics and classification. Sci Hortic (Amsterdam) 66:135–137. https://doi.org/10.1016/0304-4238(96)81078-8
Barcaccia G, Ghedina A, Lucchin M (2016) Current advances in genomics and breeding of leaf chicory (Cichorium intybus L.). Agriculture 6:50
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Teycheney PY, Geering ADW, Dasgupta I et al (2020) ICTV Virus taxonomy profile: Caulimoviridae. J Gen Virol 101:1025–1026
Verdaguer B, De Kochko A, Fux CI et al (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol Biol 37:1055–1067. https://doi.org/10.1023/A:1006004819398
Cuellar WJ, de Souza JD, Barrantes I et al (2011) Distinct cavemoviruses interact synergistically with sweet potato chlorotic stunt virus (genus crinivirus) in cultivated sweet potato. J Gen Virol 92:1233–1243. https://doi.org/10.1099/vir.0.029975-0
Pooggin MM, Fütterer J, Skryabin KG, Hohn T (1999) A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J Gen Virol 80:2217–2228. https://doi.org/10.1099/0022-1317-80-8-2217
Zheng L, Cao M, Wu L et al (2020) First identification and molecular characterization of a novel cavemovirus infecting Epiphyllum spp. Arch Virol 165:2083–2086. https://doi.org/10.1007/s00705-020-04688-2
Guindon S, Dufayard JF, Lefort V et al (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771. https://doi.org/10.1080/10635150802429642
Funding
This research was funded by Fundação de Apoio à Pesquisa do Distrito Federal, Grant no [193.001532/2016] and Conselho Nacional de Desenvolvimento Científico e Tecnológico, Grant no [305756/2017-6].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any research involving humans or animals.
Additional information
Handling Editor: Elvira Fiallo-Olivé.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
705_2021_5172_MOESM1_ESM.pdf
Supplementary file1 Supplementary Fig. S1 The results of alignment and percentage of nucelotide and amino acid sequence identity of chicory mosaic cavemovirus (ChiMV) to sweet potato collusive virus (SPCV), cassava vein mosaic virus (CsVMV), and epiphyllum virus 4 (EpV-4) (PDF 59 KB)
Rights and permissions
About this article
Cite this article
Silva, L.A., de Camargo, B.R., Eiras, M. et al. Molecular characterization of a putative new cavemovirus isolated from wild chicory (Cichorium intybus). Arch Virol 166, 2865–2868 (2021). https://doi.org/10.1007/s00705-021-05172-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05172-1