Abstract
DBR1 encodes the only known human lariat debranching enzyme and its deficiency has been found to cause an autosomal recessive inborn error of immunity characterized by pediatric brainstem viral-induced encephalitis (MIM 619441). We describe a distinct allelic disorder caused by a founder recessive DBR1 variant in four families (DBR1(NM_016216.4):c.200A > G (p.Tyr67Cys)). Consistent features include prematurity, severe intrauterine growth deficiency, congenital ichthyosis-like presentation (collodion membrane, severe skin peeling and xerosis), and death before the first year of life. Patient-derived fibroblasts displayed the characteristic accumulation of intron lariats in their RNA as revealed by targeted and untargeted analysis, in addition to a marked reduction of DBR1 on immunoblot analysis. We propose a novel DBR1-related developmental disorder that is distinct from DBR1-related encephalitis susceptibility and highlight the apparent lack of correlation with the degree of DBR1 deficiency.




Similar content being viewed by others
References
Bertoli-Avella AM, Kandaswamy KK, Khan S, Ordonez-Herrera N, Tripolszki K, Beetz C, Rocha ME, Urzi A, Hotakainen R, Leubauer A (2021) Combining exome/genome sequencing with data repository analysis reveals novel gene–disease associations for a wide range of genetic disorders. Genet Med 23:1551–1568
Chapman KB, Boeke JD (1991) Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65:483–492
Cheng J, Randall A, Baldi P (2006) Prediction of protein stability changes for single-site mutations using support vector machines. Proteins: Struct, Funct Bioinform 62:1125–1132
Choi Y, Lee H-H, Park J, Kim S, Choi S, Moon H, Shin J, Kim J-E, Choi GJ, Seo Y-S (2022) Intron turnover is essential to the development and pathogenicity of the plant pathogenic fungus Fusarium graminearum. Commun Biol 5:1129
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Ewels P, Magnusson M, Lundin S, Käller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048
Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J (2014) Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513:120–123
Graubert A, Aguet F, Ravi A, Ardlie KG, Getz G (2021) RNA-SeQC 2: Efficient RNA-seq quality control and quantification for large cohorts. Bioinformatics 37:3048–3050
Jacquier A, Rosbash M (1986) RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci 83:5835–5839
Kim J-W, Kim H-C, Kim G-M, Yang J-M, Boeke JD, Nam K (2000) Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res 28:3666–3673
Li Z, Wang S, Cheng J, Su C, Zhong S, Liu Q, Fang Y, Yu Y, Lv H, Zheng Y (2016) Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genet 12:e1006422
Li H-D, Funk CC, Price ND (2020) iREAD: a tool for intron retention detection from RNA-seq data. BMC Genomics 21:1–11
Li H-D, Lin C-X, Zheng J (2022) GTFtools: a software package for analyzing various features of gene models. Bioinformatics 38:4806–4808
Montemayor EJ, Katolik A, Clark NE, Taylor AB, Schuermann JP, Combs DJ, Johnsson R, Holloway SP, Stevens SW, Damha MJ (2014) Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1. Nucleic Acids Res 42:10845–10855
Nam K, Hudson R, Chapman KB, Ganeshan K, Damha MJ, Boeke JD (1994) Yeast lariat debranching enzyme. Substrate and sequence specificity. J Biol Chem 269:20613–20621
Nam K, Lee G, Trambley J, Devine SE, Boeke JD (1997) Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol 17:809–818
Ruskin B, Green MR (1985) An RNA processing activity that debranches RNA lariats. Science 229:135–140
Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss ME, Köster J, Marais A, Paknia O, Schröder R, Garcia-Aznar JM, Werber M (2017) Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet 25:176–182
Zhang S-Y, Casanova J-L (2015) Inborn errors underlying herpes simplex encephalitis: from TLR3 to IRF3. The Rockefeller University Press
Zhang S-Y, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, Mandel H, Garcia P, Ciancanelli MJ, Biran A (2018) Inborn errors of RNA lariat metabolism in humans with brainstem viral infection. Cell 172:952-965.e18
Zheng S, Vuong BQ, Vaidyanathan B, Lin J-Y, Huang F-T, Chaudhuri J (2015) Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell 161:762–773
Acknowledgements
We thank the study families for their enthusiastic participation.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
The referring physicians (ZR, FAM, FA, HAA) provided clinical information and images. Material preparation, data collection and analysis, and functional validation were performed by (MS, JM, MA, HES, RA-A, MR, SL and FA). PB, AB-A, FSA and all authors contributed to the writing of the manuscript, as well the final version was read and approved by all authors. FSA supervised the work in its entirety.
Corresponding author
Ethics declarations
Conflict of interest
Some authors are affiliated with Centogene, a commercial molecular laboratory.
Ethical approval
The research conformed to the principles of the Helsinki Declaration. This study was IRB-approved (KFSRHC RAC# 2121053).
Consent to participate
Informed consent was obtained for all individual participants included in the study.
Consent to publish
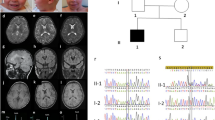
The authors affirm that human research participants provided informed consent for the publication of the images in Fig. 1.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
439_2023_2597_MOESM3_ESM.pptx
Fig S1: IGV Sashimi plot view of an intronic region in the GRK5, PDCD11, DZIP1L, TEP1, LRCH1, BRWD1 genes showing a higher intronic reads accumulation in the patient compared to the control. The low coverage in the intron-exon junction regions suggests the presence of lariat RNA. Fig S2: Panel showing expression of DBR1 in different tissues. Supplementary file3 (PPTX 571 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shamseldin, H.E., Sadagopan, M., Martini, J. et al. A founder DBR1 variant causes a lethal form of congenital ichthyosis. Hum. Genet. 142, 1491–1498 (2023). https://doi.org/10.1007/s00439-023-02597-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-023-02597-3