Abstract
Objectives
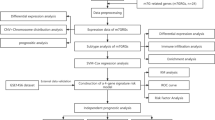
This study aims to develop and validate a prognostic signature based on 7-methylguanosine-related (M7G-related) miRNAs for predicting prognosis and immune implications in breast invasive carcinoma (BRCA).
Materials and methods
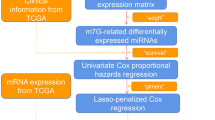
M7G-related miRNA data of BRCA were obtained from The Cancer Genome Atlas (TCGA). Least absolute shrinkage and selection operator (LASSO)-penalized, univariate, and multivariate Cox regression analyses were used to construct the prognostic signature. Furthermore, the predictive validity was verified using Kaplan–Meier (KM) survival risk and receiver operating characteristic (ROC) plots. Internal random sampling verification was used to simplify and validate the signature. RT-qPCR was used to quantify the expression level of transcriptional profiles. The independent prognostic role of the risk score was validated using univariate and multivariate regression. Single-sample Gene Set Enrichment Analysis (ssGSEA) was used for functional and immune enrichment analysis.
Results
A total of 18 M7G-related miRNAs were identified to construct the prognostic signature in BRCA. The low-risk group exhibited significantly higher overall survival than the high-risk group in the KM survival plot (P < 0.001). The area under the curve (AUC) for 1-, 3-, and 5-year survivals in the ROC curve were 0.737, 0.724, and 0.702, respectively. The survival significance in the training and testing cohorts was confirmed by random sampling verification. The most prominent miRNAs in the signature were the miR-7, miR-139, miR-10b, and miR-4728. Furthermore, immune scores for B, mast, and Th1 cells varied between risk groups. Our research demonstrated that CD52 was the most positively correlated gene with immune cells and functions in BRCA.
Conclusion
Our study presents a comprehensive and systematic analysis of M7G-related miRNAs to construct a prognostic signature in BRCA. The signature demonstrated excellent prognostic validity, with the risk score as an independent prognostic factor. These results provide critical evidence for further investigation of M7G miRNAs and offer new insights for BRCA patients in the context of effective immunotherapy.














Similar content being viewed by others
Data availability
TCGA: https://portal.gdc.cancer.gov/repository; TargetScan: https://www.targetscan.org/vert_80/; Gencode: https://www.gencodegenes.org/; miRbase: https://www.mirbase.org/; MDAnderson bioinformatics center: https://bioinformatics.mdanderson.org/estimate/disease.html; KEGG: https://www.genome.jp/kegg/; GO: http://geneontology.org/docs/go-enrichment-analysis/.
Abbreviations
- M7G:
-
7-Methylguanosine
- miRNA:
-
MicroRNA
- BRCA:
-
Breast invasive carcinoma
- TCGA:
-
The cancer genome atlas
- LASSO:
-
Least absolute shrinkage and selection operator
- KM:
-
Kaplan–Meier
- ROC:
-
Receiver operating characteristic
- ssGSEA:
-
Single sample gene set enrichment analysis
- AUC:
-
Area under curve
- BC:
-
Breast cancer
- METTL1:
-
Methyltransferase like 1
- WDR4:
-
WD repeat domain 4
- HCC:
-
Hepatocellular carcinoma
- CAFs:
-
Cancer associated fibroblasts
- TME:
-
Tumor microenvironment
- EMT:
-
Epithelial-mesenchymal transition
- MRBCmiRs:
-
M7G related breast invasive carcinoma miRNAs
- MRDEmiRs:
-
M7G related different expression miRNAs
- DERRmRs:
-
Different expressed risk-score related mRNAs
- DEIRmRs:
-
Different expressed immune-score related mRNAs
- PCA:
-
Principal component analysis
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- DO:
-
Disease ontology
- GO:
-
Gene ontology
References
Aponte-López A, Fuentes-Pananá EM, Cortes-Muñoz D, Muñoz-Cruz S (2018) Mast cell, the neglected member of the tumor microenvironment: role in breast cancer. J Immunol Res 2018:2584243. https://doi.org/10.1155/2018/2584243
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Bertoli G, Cava C, Castiglioni I (2015) MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 5:1122–1143. https://doi.org/10.7150/thno.11543
Biagioni F et al (2013) The locus of microRNA-10b: a critical target for breast cancer insurgence and dissemination. Cell Cycle 12:2371–2375. https://doi.org/10.4161/cc.25380
Boccaletto P et al (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. https://doi.org/10.1093/nar/gkx1030
Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23:175–205. https://doi.org/10.1146/annurev.cellbio.23.090506.123406
Canu V et al (2021) Aberrant transcriptional and post-transcriptional regulation of SPAG5, a YAP-TAZ-TEAD downstream effector, fuels breast cancer cell proliferation. Cell Death Differ 28:1493–1511. https://doi.org/10.1038/s41418-020-00677-9
Cao P et al (2022) hsa_circ_0003410 promotes hepatocellular carcinoma progression by increasing the ratio of M2/M1 macrophages through the miR-139-3p/CCL5 axis. Cancer Sci 113:634–647. https://doi.org/10.1111/cas.15238
Cui YX et al (2017) MicroRNA-7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer cells. Br J Cancer 117:89–101. https://doi.org/10.1038/bjc.2017.156
Dai G et al (2018) Colorectal cancer cell-derived exosomes containing miR-10b regulate fibroblast cells via the PI3K/Akt pathway. Bull Cancer 105:336–349. https://doi.org/10.1016/j.bulcan.2017.12.009
Deblois G et al (2020) Epigenetic switch-induced viral mimicry evasion in chemotherapy-resistant breast cancer. Cancer Discov 10:1312–1329. https://doi.org/10.1158/2159-8290.Cd-19-1493
DeSantis CE et al (2019) Breast cancer statistics, 2019. CA Cancer J Clin 69:438–451. https://doi.org/10.3322/caac.21583
Erbes T et al (2016) Hyperthermia-driven aberrations of secreted microRNAs in breast cancer in vitro. Int J Hyperthermia 32:630–642. https://doi.org/10.3109/02656736.2016.1161832
Fang F et al (2015) MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol 63:874–885. https://doi.org/10.1016/j.jhep.2015.05.008
Fedoriw A et al (2022) Inhibiting type I arginine methyltransferase activity promotes T cell-mediated antitumor immune responses. Cancer Immunol Res 10:420–436. https://doi.org/10.1158/2326-6066.Cir-21-0614
Fei S, Cao L, Li S (2021) microRNA-139-5p alleviates neurological deficit in hypoxic-ischemic brain damage via HDAC4 depletion and BCL-2 activation. Brain Res Bull 169:73–80. https://doi.org/10.1016/j.brainresbull.2020.12.020
Garaud S et al (2019) Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight. https://doi.org/10.1172/jci.insight.129641
Garcia-Estevez L, Moreno-Bueno G (2019) Updating the role of obesity and cholesterol in breast cancer. Breast Cancer Res 21:35. https://doi.org/10.1186/s13058-019-1124-1
Garcia-Martinez L, Zhang Y, Nakata Y, Chan HL, Morey L (2021) Epigenetic mechanisms in breast cancer therapy and resistance. Nat Commun 12:1786. https://doi.org/10.1038/s41467-021-22024-3
Gevers Leuven JA (1994) Sex steroids and lipoprotein metabolism. Pharmacol Ther 64:99–126. https://doi.org/10.1016/0163-7258(94)90035-3
Gu-Trantien C et al (2013) CD4+ follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest 123:2873–2892. https://doi.org/10.1172/jci67428
Guy MP, Phizicky EM (2014) Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol 11:1608–1618. https://doi.org/10.1080/15476286.2015.1008360
Hong T, Ding J, Li W (2019) miR-7 reverses breast cancer resistance to chemotherapy by targeting MRP1 and BCL2. Onco Targets Ther 12:11097–11105. https://doi.org/10.2147/ott.S213780
Hu L et al (2021) MicroRNA-7 overexpression positively regulates the CD8(+) SP cell development via targeting PIK3R1. Exp Cell Res 407:112824. https://doi.org/10.1016/j.yexcr.2021.112824
Huynh FC, Jones FE (2014) MicroRNA-7 inhibits multiple oncogenic pathways to suppress HER2Δ16 mediated breast tumorigenesis and reverse trastuzumab resistance. PloS One 9:e114419. https://doi.org/10.1371/journal.pone.0114419
Iorio MV et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070. https://doi.org/10.1158/0008-5472.Can-05-1783
Jiang Y, Jiang J, Jia H, Qiao Z, Zhang J (2018) Recovery of miR-139-5p in ovarian cancer reverses cisplatin resistance by targeting C-Jun. Cell Physiol Biochem 51:129–141. https://doi.org/10.1159/000495169
Kastl L, Brown I, Schofield AC (2012) miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat 131:445–454. https://doi.org/10.1007/s10549-011-1424-3
Khalili N et al (2020) Diagnostic, prognostic, and therapeutic significance of miR-139–5p in cancers. Life Sci 256:117865. https://doi.org/10.1016/j.lfs.2020.117865
Li Q et al (2016) miR-139-5p inhibits the epithelial-mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep 6:27157. https://doi.org/10.1038/srep27157
Li B, Yang C, Zhu Z, Chen H, Qi B (2022) Hypoxic glioma-derived extracellular vesicles harboring MicroRNA-10b-5p enhance M2 polarization of macrophages to promote the development of glioma. CNS Neurosci Ther 28:1733–1747. https://doi.org/10.1111/cns.13905
Lin S et al (2018a) Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell 71:244-255.e245. https://doi.org/10.1016/j.molcel.2018.06.001
Lin H et al (2018b) Protein arginine methyltransferase 8 gene enhances the colon cancer stem cell (CSC) function by upregulating the pluripotency transcription factor. J Cancer 9:1394–1402. https://doi.org/10.7150/jca.23835
Liu H et al (2016) miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract 212:643–649. https://doi.org/10.1016/j.prp.2016.04.011
Liu W et al (2017) A systems biology approach to identify microRNAs contributing to cisplatin resistance in human ovarian cancer cells. Mol Biosyst 13:2268–2276. https://doi.org/10.1039/c7mb00362e
Liu Y, Yang Y, Du J, Lin D, Li F (2020) MiR-3613-3p from carcinoma-associated fibroblasts exosomes promoted breast cancer cell proliferation and metastasis by regulating SOCS2 expression. IUBMB Life 72:1705–1714. https://doi.org/10.1002/iub.2292
Ma J et al (2021a) METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther 29:3422–3435. https://doi.org/10.1016/j.ymthe.2021.08.005
Ma YF et al (2021b) The immune-related gene CD52 is a favorable biomarker for breast cancer prognosis. Gland Surg 10:780–798. https://doi.org/10.21037/gs-20-922
Masuda M et al (2012) An induction of microRNA, miR-7 through estrogen treatment in breast carcinoma. J Transl Med. https://doi.org/10.1186/1479-5876-10-s1-s2
Orellana EA et al (2021) METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell 81:3323-3338.e3314. https://doi.org/10.1016/j.molcel.2021.06.031
Pandolfini L et al (2019) METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol Cell 74:1278-1290.e1279. https://doi.org/10.1016/j.molcel.2019.03.040
Pang RW, Lam KS (2002) Hormonal influences on lipoprotein(a) metabolism. Diabetes Obes Metab 4:156–165. https://doi.org/10.1046/j.1463-1326.2002.00176.x
Pencheva N, Tavazoie SF (2013) Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol 15:546–554. https://doi.org/10.1038/ncb2769
Persson H et al (2011) Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res 71:78–86. https://doi.org/10.1158/0008-5472.Can-10-1869
Pogribny IP et al (2010) Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 127:1785–1794. https://doi.org/10.1002/ijc.25191
Seong M, Lee J, Kang H (2019) Hypoxia-induced regulation of mTOR signaling by miR-7 targeting REDD1. J Cell Biochem 120:4523–4532. https://doi.org/10.1002/jcb.27740
Shao Q et al (2018) MicroRNA-139-5p affects cisplatin sensitivity in human nasopharyngeal carcinoma cells by regulating the epithelial-to-mesenchymal transition. Gene 652:48–58. https://doi.org/10.1016/j.gene.2018.02.003
Shao YY et al (2022) Low miR-10b-3p associated with sorafenib resistance in hepatocellular carcinoma. Br J Cancer 126:1806–1814. https://doi.org/10.1038/s41416-022-01759-w
Stadler SC, Hacker U, Burkhardt R (2016) Cholesterol metabolism and breast cancer. Curr Opin Lipidol 27:200–201. https://doi.org/10.1097/mol.0000000000000285
Su T et al (2022) miR-7/TGF-β2 axis sustains acidic tumor microenvironment-induced lung cancer metastasis. Acta Pharm Sin B 12:821–837. https://doi.org/10.1016/j.apsb.2021.06.009
Sung H et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Trifari S et al (2013) MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci U S A 110:18608–18613. https://doi.org/10.1073/pnas.1317191110
Tu J et al (2022) MicroRNA-10b promotes arthritis development by disrupting CD4(+) T cell subtypes. Mol Ther Nucleic Acids 27:733–750. https://doi.org/10.1016/j.omtn.2021.12.022
Wang Y et al (2017) MiR-139-5p protect against myocardial ischemia and reperfusion (I/R) injury by targeting autophagy-related 4D and inhibiting AMPK/mTOR/ULK1 pathway. Int J Clin Exp Pathol 10:10140–10151
Wang J et al (2020) CD52 is a prognostic biomarker and associated with tumor microenvironment in breast cancer. Front Genet 11:578002. https://doi.org/10.3389/fgene.2020.578002
Xia P et al (2021) MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis 12:691. https://doi.org/10.1038/s41419-021-03973-5
Xu G et al (2019) Exosomal miRNA-139 in cancer-associated fibroblasts inhibits gastric cancer progression by repressing MMP11 expression. Int J Biol Sci 15:2320–2329. https://doi.org/10.7150/ijbs.33750
Yager JD, Davidson NE (2006) Estrogen carcinogenesis in breast cancer. N Engl J Med 354:270–282. https://doi.org/10.1056/NEJMra050776
Yang L, Yin D, Wang Y, Cao L (2017) Inhibition of the growth of hepatocellular carcinoma cells through fibroblast growth factor 18 suppressed by miR-139. Oncol Rep 38:2565–2571. https://doi.org/10.3892/or.2017.5869
Zhang HD et al (2015) MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun 465:702–713. https://doi.org/10.1016/j.bbrc.2015.08.053
Zhang LS et al (2019) Transcriptome-wide mapping of internal N(7)-methylguanosine methylome in mammalian mRNA. Mol Cell 74:1304-1316.e1308. https://doi.org/10.1016/j.molcel.2019.03.036
Zhang X, Gu G, Li X, Zhang C (2020) Lidocaine alleviates cisplatin resistance and inhibits migration of MGC-803/DDP cells through decreasing miR-10b. Cell Cycle 19:2530–2537. https://doi.org/10.1080/15384101.2020.1809914
Zhao J et al (2021) C/EBPα/miR-7 controls CD4(+) T-cell activation and function and orchestrates experimental autoimmune hepatitis in mice. Hepatology 74:379–396. https://doi.org/10.1002/hep.31607
Zhao Y et al (2021) m7G methyltransferase METTL1 promotes post-ischemic angiogenesis via promoting VEGFA mRNA translation. Front Cell Dev Biol 9:642080. https://doi.org/10.3389/fcell.2021.642080
Zheng NN et al (2020) Combining protein arginine methyltransferase inhibitor and anti-programmed death-ligand-1 inhibits pancreatic cancer progression. World J Gastroenterol 26:3737–3749. https://doi.org/10.3748/wjg.v26.i26.3737
Zhou Y et al (2021) HER2-intronic miR-4728–5p facilitates HER2 expression and accelerates cell proliferation and migration by targeting EBP1 in breast cancer. PloS One 16:e0245832. https://doi.org/10.1371/journal.pone.0245832
Acknowledgements
We appreciate the databases of TCGA, Targetscan, Gencode, miRbase, MDAnderson bioinformatics center, KEGG and GO for the availability of the data.
Funding
This article was funded by the Fundamental Research Funds for the Central Universities of Central South University (Grant Nos.2021zzts0371). The funding doesn’t influence study design, collection, analysis and interpretation of data; the writing of the report; and the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
AYL, TX and HL: Conceptualization, Formal analysis. AYL and HNX: Writing original draft. AYL, TX and HL: Review, Editing. AYL, ZYZ, CX, YX, BY, PXW, and ZYC: Visualization, Methodology. TX and HL: Supervision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, AY., Xiao, HN., Zhao, ZY. et al. Prognostic and immune implications of a novel 7-methylguanosine-related microRNA signature in breast invasive carcinoma: from exploration to validation. J Cancer Res Clin Oncol 149, 9105–9128 (2023). https://doi.org/10.1007/s00432-023-04849-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04849-1