Abstract
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disorder. It is mostly sporadic, with the C9orf72 repeat expansion being the most common genetic cause. While the prevalence of C9orf72-ALS in patients from different populations has been studied, data regarding the yield of C9orf72 compared to an ALS gene panel testing is limited.
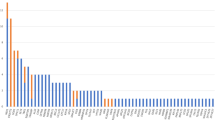
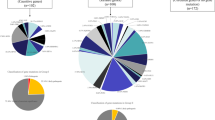
We aimed to explore the application of C9orf72 versus a gene panel in the general Israeli population. A total of 140 ALS patients attended our Neurogenetics Clinic throughout 2018–2023. Disease onset was between ages 60 and 69 years for most patients (34%); however, a quarter had an early-onset disease (< 50 years). Overall, 119 patients (85%) were genetically evaluated: 116 (97%) were tested for the C9orf72 repeat expansion and 64 (54%) underwent gene panel testing. The C9orf72 repeat expansion had a prevalence of 21% among Ashkenazi Jewish patients compared to 5.7% in non-Ashkenazi patients, while the gene panel had a higher yield in non-Ashkenazi patients with 14% disease-causing variants compared to 5.7% in Ashkenazi Jews. Among early-onset ALS patients, panel testing was positive in 12% compared to 2.9% for C9orf72.
We suggest a testing strategy for the Israeli ALS patients: C9orf72 should be the first-tier test in Ashkenazi Jewish patients, while a gene panel should be considered as the first step in non-Ashkenazi and early-onset patients. Tiered testing has important implications for patient management, including prognosis, ongoing clinical trials, and prevention in future generations. Similar studies should be implemented worldwide to uncover the diverse ALS genetic architecture and facilitate tailored care.



Similar content being viewed by others
Data availability
Data related to the study are available from the corresponding authors upon reasonable request.
References
Hardiman O, Al-Chalabi A, Chio A et al (2017) Amyotrophic lateral sclerosis. Nat Rev Dis Prim 3:17071
Akçimen F, Lopez ER, Landers JE et al (2023) Amyotrophic lateral sclerosis: translating genetic discoveries into therapies. Nat Rev Genet 24:642–658. https://doi.org/10.1038/s41576-023-00592-y
Su XW, Broach JR, Connor JR et al (2014) Genetic heterogeneity of amyotrophic lateral sclerosis: implications for clinical practice and research. Muscle Nerve 49:786–803. https://doi.org/10.1002/mus.24198
Majounie E, Renton AE, Mok K et al (2012) Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol 11:323–330. https://doi.org/10.1016/S1474-4422(12)70043-1
Dejesus-hernandez M, Mackenzie IR, Boeve BF et al (2012) Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis Mariely. Neuron 72:245–256. https://doi.org/10.1016/j.neuron.2011.09.011.Expanded
Murphy NA, Arthur KC, Tienari PJ et al (2017) Age-related penetrance of the C9orf72 repeat expansion. Sci Rep 7:1–7. https://doi.org/10.1038/s41598-017-02364-1
Pensato V, Magri S, Bella ED et al (2020) Sorting rare ALS genetic variants by targeted re-sequencing panel in italian patients: OPTN, VCP, and SQSTM1 variants account for 3% of rare genetic forms. J Clin Med 9:1–18. https://doi.org/10.3390/jcm9020412
Mesaros M, Lenz S, Lim W et al (2022) Investigating the Genetic Profile of the Amyotrophic Lateral Sclerosis/Frontotemporal Dementia (ALS-FTD) Continuum in Patients of Diverse Race, Ethnicity and Ancestry. Genes (Basel). https://doi.org/10.3390/genes13010076
Laaksovirta H, Launes J, Jansson L et al (2022) ALS in Finland: major genetic variants and clinical characteristics of patients with and without the C9orf72 hexanucleotide repeat expansion. Neurol Genet. https://doi.org/10.1212/NXG.0000000000000665
Orrù S, Manolakos E, Orrù N et al (2012) High frequency of the TARDBP p.Ala382Thr mutation in Sardinian patients with amyotrophic lateral sclerosis. Clin Genet 81:172–178. https://doi.org/10.1111/j.1399-0004.2011.01668.x
Zlotogora J, van Baal S, Patrinos GP (2009) The Israeli National Genetic Database. IMAJ 11:373–375
Goldstein O, Gana-Weisz M, Nefussy B et al (2018) High frequency of C9orf72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis patients from two founder populations sharing the same risk haplotype. Neurobiol Aging 64:160.e1-160.e7. https://doi.org/10.1016/j.neurobiolaging.2017.12.015
Bourinaris T, Houlden H (2018) C9orf72 and its relevance in parkinsonism and movement disorders: a comprehensive review of the literature. Mov Disord Clin Pract 5:575–585. https://doi.org/10.1002/mdc3.12677
González-Pérez P, Cirulli ET, Drory VE et al (2012) Novel mutation in VCP gene causes atypical amyotrophic lateral sclerosis. Neurology 79:2201–2208. https://doi.org/10.1212/WNL.0b013e318275963b
Goldstein O, Nayshool O, Nefussy B et al (2016) OPTN 691–692insAG is a founder mutation causing recessive ALS and increased risk in heterozygotes. Neurology 86:446–453. https://doi.org/10.1212/WNL.0000000000002334
Van Daele SH, Moisse M, van Vugt JJFA et al (2023) Genetic variability in sporadic amyotrophic lateral sclerosis. Brain 146:3760–3769. https://doi.org/10.1093/brain/awad120
Maruyama H, Morino H, Ito H et al (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465:223–226. https://doi.org/10.1038/nature08971
Miller TM, Cudkowicz ME, Genge A et al (2022) Trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med 387:1099–1110. https://doi.org/10.1056/nejmoa2204705
Corrado L, D’Alfonso S, Bergamaschi L et al (2006) SOD1 gene mutations in Italian patients with Sporadic Amyotrophic Lateral Sclerosis (ALS). Neuromuscul Disord 16:800–804. https://doi.org/10.1016/j.nmd.2006.07.004
Gellera C, Castellotti B, Riggio MC et al (2001) Superoxide dismutase gene mutations in Italian patients with familial and sporadic amyotrophic lateral sclerosis: Identification of three novel missense mutations. Neuromuscul Disord 11:404–410. https://doi.org/10.1016/S0960-8966(00)00215-7
Fujisawa T, Homma K, Yamaguchi N et al (2012) A novel monoclonal antibody reveals a conformational alteration shared by amyotrophic lateral sclerosis-linked SOD1 mutants. Ann Neurol 72:739–749. https://doi.org/10.1002/ana.23668
Vucic S, Rothstein JD, Kiernan MC (2014) Advances in treating amyotrophic lateral sclerosis: Insights from pathophysiological studies. Trends Neurosci 37:433–442. https://doi.org/10.1016/j.tins.2014.05.006
Suzuki N, Nishiyama A, Warita H, Aoki M (2023) Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy. J Hum Genet 68:131–152. https://doi.org/10.1038/s10038-022-01055-8
Dolzhenko E, Deshpande V, Schlesinger F et al (2019) ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics 35:4754–4756. https://doi.org/10.1093/bioinformatics/btz431
Acknowledgements
We thank the patients and their families, and acknowledge the Molecular Genetics Laboratory and the Genomics Center staff at the Tel Aviv Sourasky Medical Center for performing the genetic workup.
Funding
None.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest.
Ethical approval
Genetic testing was performed as a clinical service under standard clinical consent forms. The study was approved by the Tel Aviv Sourasky Medical Center Institutional Helsinki Committee (#0586–20-TLV), which grants a waiver of informed consent for retrospective study of patient medical records.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Barel, D., Marom, D., Ponger, P. et al. Genetic diagnosis and detection rates using C9orf72 repeat expansion and a multi-gene panel in amyotrophic lateral sclerosis. J Neurol (2024). https://doi.org/10.1007/s00415-024-12368-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00415-024-12368-3