Abstract
Background
Phenotypic heterogeneity within or between families with a same deep-intronic splice-altering variant in the DMD gene has never been systematically analyzed. This study aimed to determine the phenotypic and genetic characteristics of patients with deep-intronic DMD variants.
Methods
Of 1338 male patients with a suspected dystrophinopathy, 38 were confirmed to have atypical pathogenic DMD variants via our comprehensive genetic testing approach. Of the 38 patients, 30 patients from 22 unrelated families with deep-intronic DMD variants underwent a detailed clinical and imaging assessment.
Results
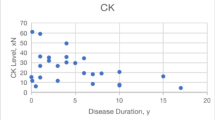
Nineteen different deep-intronic DMD variants were identified in the 30 patients, including 15 with Duchenne muscular dystrophy (DMD), 14 with Becker muscular dystrophy (BMD), and one with X-linked dilated cardiomyopathy. Of the 19 variants, 15 were single-nucleotide variants, 2 were structural variants (SVs), and 2 were pure-intronic large-scale SVs causing aberrant inclusion of other protein-coding genes sequences into the mature DMD transcripts. The trefoil with single fruit sign was observed in 18 patients and the concentric fatty infiltration pattern was observed in 2 patients. Remarkable phenotypic heterogeneity was observed not only in skeletal but also cardiac muscle involvement in 2 families harboring a same deep-intronic variant. Different skeletal muscle involvement between families with a same variant was observed in 4 families. High inter-individual phenotypic heterogeneity was observed within two BMD families and one DMD family.
Conclusions
Our study first highlights the variable phenotypic expressivity of deep-intronic DMD variants and demonstrates a new class of deep-intronic DMD variants, i.e., pure-intronic SVs involving other protein-coding genes.





Similar content being viewed by others
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Information.
References
Muntoni F, Torelli S, Ferlini A (2003) Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol 2:731–740
Xie Z et al (2021) Practical approach to the genetic diagnosis of unsolved dystrophinopathies: a stepwise strategy in the genomic era. J Med Genet 58:743–751
Xie Z et al (2020) Long-read whole-genome sequencing for the genetic diagnosis of dystrophinopathies. Ann Clin Transl Neurol 7:2041–2046
Waddell LB et al (2021) WGS and RNA studies diagnose noncoding DMD variants in males with high creatine kinase. Neurol Genet 7:e554
de Feraudy Y et al (2021) Very low residual dystrophin quantity is associated with milder dystrophinopathy. Ann Neurol 89:280–292
Tong YR et al (2020) a comprehensive analysis of 2013 dystrophinopathies in China: a report from National Rare Disease Center. Front Neurol 11:572006
Neri M et al (2020) The genetic landscape of dystrophin mutations in Italy: a nationwide study. Front Genet 11:131
Xie Z et al (2020) Splicing characteristics of dystrophin pseudoexons and identification of a novel pathogenic intronic variant in the DMD gene. Genes (Basel) 11:1180
Bladen CL et al (2015) The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat 36:395–402
Fratter C et al (2020) EMQN best practice guidelines for genetic testing in dystrophinopathies. Eur J Hum Genet 28:1141–1159
Guo R et al (2015) DMD mutation spectrum analysis in 613 Chinese patients with dystrophinopathy. J Hum Genet 60:435–442
Okubo M et al (2017) Comprehensive analysis for genetic diagnosis of Dystrophinopathies in Japan. Orphanet J Rare Dis 12:149
Juan-Mateu J et al (2013) Interplay between DMD point mutations and splicing signals in Dystrophinopathy phenotypes. PLoS ONE 8:e59916
Jones HF et al (2019) Importance of muscle biopsy to establish pathogenicity of DMD missense and splice variants. Neuromuscul Disord 29:913–919
Cummings BB et al (2017) Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med 9
Lu X et al (2021) Novel intronic mutations introduce pseudoexons in DMD that cause muscular dystrophy in patients. Front Genet 12:657040
Waldrop MA et al (2022) Intron mutations and early transcription termination in Duchenne and Becker muscular dystrophy. Hum Mutat 43:511–528
Keegan NP, Wilton SD, Fletcher S (2021) Analysis of pathogenic pseudoexons reveals novel mechanisms driving cryptic splicing. Front Genet 12:806946
Falzarano MS et al (2022) RNA-seq in DMD urinary stem cells recognized muscle-related transcription signatures and addressed the identification of atypical mutations by whole-genome sequencing. HGG Adv 3:100054
Xie Z et al (2019) Value of muscle magnetic resonance imaging in the differential diagnosis of muscular dystrophies related to the dystrophin-glycoprotein complex. Orphanet J Rare Dis 14:250
Richards S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Scott OM, Hyde SA, Goddard C, Dubowitz V (1982) Quantitation of muscle function in children: a prospective study in Duchenne muscular dystrophy. Muscle Nerve 5:291–301
Ling C et al (2020) Exonic rearrangements in DMD in Chinese Han individuals affected with Duchenne and Becker muscular dystrophies. Hum Mutat 41:668–677
Tuffery-Giraud S et al (2009) Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat 30:934–945
Burset M, Seledtsov IA, Solovyev VV (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res 29:255–259
Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C (2009) Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67
Anna A, Monika G (2018) Splicing mutations in human genetic disorders: examples, detection, and confirmation. J Appl Genet 59:253–268
Tuffery-Giraud S, Miro J, Koenig M, Claustres M (2017) Normal and altered pre-mRNA processing in the DMD gene. Hum Genet 136:1155–1172
Waldrop MA et al (2020) Clinical phenotypes of DMD exon 51 skip equivalent deletions: a systematic review. J Neuromuscul Dis 7:217–229
Coimbra Neto AR et al (2021) Intrafamilial phenotypic heterogeneity related to a new DMD splice site variant. Neuromuscul Disord 31:788–797
Zatz M et al (2014) Milder course in Duchenne patients with nonsense mutations and no muscle dystrophin. Neuromuscul Disord 24:986–989
Gao QQ, McNally EM (2015) The dystrophin complex: structure, function, and implications for therapy. Compr Physiol 5:1223–1239
Funding
This study was supported by the Beijing Municipal Science and Technology Commission (Grant number Z191100006619034) and National Natural Science Foundation of China (Grant number 82201553).
Author information
Authors and Affiliations
Contributions
ZX, CS, CL, ZX, SML, IS, ZW and YY contributed to the study conception and design. Material preparation and data collection were performed by ZX, CS, CL, JY, CL, XG, YL, MY, YL, LW, LM, YS and JD. Clinical and imaging data analysis was performed by ZX, CS, CL, JY, LW, LM, YS, JD, ZW and YY. Genetic data analysis was performed by ZX, CS, CL, ZX, ZW and YY. Bioinformatic analysis was performed by ZX, CS, ZX, SML and IS. Figure layout was performed by ZX, CS, CL, SML, IS, ZW and YY. The first draft of the manuscript was written by ZX and CS, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Ethics statement
This study was approved by the Ethics Committee at Peking University First Hospital (2019181). Informed consent was obtained from adult patients or parents/guardians of minors included in the study cohort.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, Z., Sun, C., Liu, C. et al. Clinical, muscle imaging, and genetic characteristics of dystrophinopathies with deep-intronic DMD variants. J Neurol 270, 925–937 (2023). https://doi.org/10.1007/s00415-022-11432-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-022-11432-0