Abstract
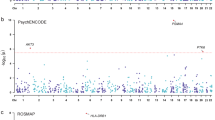
The establishment of 3ʹaQTLs comprehensive database provides an opportunity to help explore the functional interpretation from the genome-wide association study (GWAS) data of psychiatric disorders. In this study, we aim to search novel susceptibility genes, pathways, and related chemicals of five psychiatric disorders via GWAS and 3ʹaQTLs datasets. The GWAS datasets of five psychiatric disorders were collected from the open platform of Psychiatric Genomics Consortium (PGC, https://www.med.unc.edu/pgc/) and iPSYCH (https://ipsych.dk/) (Demontis et al. in Nat Genet 51(1):63–75, 2019; Grove et al. in Nat Genet 51:431–444, 2019; Genomic Dissection of Bipolar Disorder and Schizophrenia in Cell 173: 1705-1715.e1716, 2018; Mullins et al. in Nat Genet 53: 817–829; Howard et al. in Nat Neurosci 22: 343–352, 2019). The 3′untranslated region (3′UTR) alternative polyadenylation (APA) quantitative trait loci (3′aQTLs) summary datasets of 12 brain regions were obtained from another public platform (https://wlcb.oit.uci.edu/3aQTLatlas/) (Cui et al. in Nucleic Acids Res 50: D39–D45, 2022). First, we aligned the GWAS-associated SNPs of psychiatric disorders and datasets of 3′aQTLs, and then, the GWAS-associated 3ʹaQTLs were identified from the overlap. Second, gene ontology (GO) and pathway analysis was applied to investigate the potential biological functions of matching genes based on the methods provided by MAGMA. Finally, chemical-related gene-set analysis (GSA) was also conducted by MAGMA to explore the potential interaction of GWAS-associated 3ʹaQTLs and multiple chemicals in the mechanism of psychiatric disorders. A number of susceptibility genes with 3ʹaQTLs were found to be associated with psychiatric disorders and some of them had brain-region specificity. For schizophrenia (SCZ), HLA-A showed associated with psychiatric disorders in all 12 brain regions, such as cerebellar hemisphere (P = 1.58 × 10–36) and cortex (P = 1.58 × 10–36). GO and pathway analysis identified several associated pathways, such as Phenylpropanoid Metabolic Process (GO:0009698, P = 6.24 × 10–7 for SCZ). Chemical-related GSA detected several chemical-related gene sets associated with psychiatric disorders. For example, gene sets of Ferulic Acid (P = 6.24 × 10–7), Morin (P = 4.47 × 10–2) and Vanillic Acid (P = 6.24 × 10–7) were found to be associated with SCZ. By integrating the functional information from 3ʹaQTLs, we identified several susceptibility genes and associated pathways especially chemical-related gene sets for five psychiatric disorders. Our results provided new insights to understand the etiology and mechanism of psychiatric disorders.


Similar content being viewed by others
Data availability
The UK Biobank data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. We will return the derived data fields following UK Biobank policy; in due course, they will be available through the UK Biobank Access Management System. The CTD are available through the CTD Access Management System https://ctdbase.org/. We will return the derived data fields following CTD policy; in due course, they will be available through the CTD Access Management System.
References
Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E et al (2019) Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51(1):63–75
Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H et al (2019) Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51(3):431–444
Genomic Dissection of Bipolar Disorder and Schizophrenia (2018) Including 28 Subphenotypes. Cell 173(7):1705-1715.e1716
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z et al (2021) Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet 53(6):817–829
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M et al (2019) Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22(3):343–352
Cui Y, Peng F, Wang D, Li Y, Li JS, Li L et al (2022) 3’aQTL-atlas: an atlas of 3’UTR alternative polyadenylation quantitative trait loci across human normal tissues. Nucleic Acids Res 50(D1):D39–D45
Battle DE (2013) Diagnostic and Statistical Manual of Mental Disorders (DSM). Codas 25(2):191–192
Costello EJ, Egger H, Angold A (2005) 10-Year research update review: the epidemiology of child and adolescent psychiatric disorders: I. Methods and public health burden. J Am Acad Child Adolesc Psychiatry 44(10):972–986
Saha S, Chant D, Welham J, McGrath J (2005) A systematic review of the prevalence of schizophrenia. PLoS Med 2(5):e141
Gonda X, Petschner P, Eszlari N, Baksa D, Edes A, Antal P et al (2019) Genetic variants in major depressive disorder: From pathophysiology to therapy. Pharmacol Ther 194:22–43
Kessler RC, Bromet EJ The epidemiology of depression across cultures. (1545–2093 (Electronic)).
Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA (2007) The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am J Psychiatry 164(6):942–948
Holt R, Barnby G, Maestrini E, Bacchelli E, Brocklebank D, Sousa I et al (2010) Linkage and candidate gene studies of autism spectrum disorders in European populations. Eur J Human Genet EJHG 18(9):1013–1019
Baird G, Simonoff E, Pickles A, Chandler S, Loucas T, Meldrum D et al (2006) Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP). The Lancet 368(9531):210–215
Müller JK, Leweke FM (2016) Bipolar disorder: clinical overview. Med Monatsschr Pharm 39(9):363–369
Plomin R, Owen MJ, McGuffin P (1994) The genetic basis of complex human behaviors. Science 264(5166):1733–1739
Arango C, Dragioti E, Solmi M, Cortese S, Domschke K, Murray RM et al (2021) Risk and protective factors for mental disorders beyond genetics: an evidence-based atlas. World Psychiatry 20(3):417–436
Eubig PA, Aguiar A, Schantz SL (2010) Lead and PCBs as risk factors for attention deficit/hyperactivity disorder. Environ Health Perspect 118(12):1654–1667
Keil KP, Lein PJ. DNA methylation: a mechanism linking environmental chemical exposures to risk of autism spectrum disorders? Environ Epigenet 2016; 2(1).
Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF et al (2009) Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. The Lancet 373(9659):234–239
Grimm O, Kranz TM, Reif A (2020) Genetics of ADHD: what should the clinician know? Curr Psychiatry Rep 22(4):18–18
Colvert E, Tick B, McEwen F, Stewart C, Curran SR, Woodhouse E et al (2015) Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiat 72(5):415–423
Cheng S, Guan F, Ma M, Zhang L, Cheng B, Qi X et al (2020) An atlas of genetic correlations between psychiatric disorders and human blood plasma proteome. Eur Psychiatry 63(1):e17
Zheng Z-H, Tu J-L, Li X-H, Hua Q, Liu W-Z, Liu Y et al (2021) Neuroinflammation induces anxiety- and depressive-like behavior by modulating neuronal plasticity in the basolateral amygdala. Brain Behav Immun 91:505–518
Konarski JZ, McIntyre RS, Grupp LA, Kennedy SH (2005) Is the cerebellum relevant in the circuitry of neuropsychiatric disorders? J Psychiatry Neurosci JPN 30(3):178–186
Ellison-Wright I, Bullmore E (2010) Anatomy of bipolar disorder and schizophrenia: a meta-analysis. Schizophr Res 117(1):1–12
Li L, Huang K-L, Gao Y, Cui Y, Wang G, Elrod ND et al (2021) An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability. Nat Genet 53(7):994–1005
Sheng G, dos Reis M, Stern CD (2003) Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell 115(5):603–613
Human genomics (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348(6235):648–660
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Lindenbaum P, Redon R (2018) bioalcidae, samjs and vcffilterjs: object-oriented formatters and filters for bioinformatics files. Bioinformatics 34(7):1224–1225
Shabalin AA (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28(10):1353–1358
Stegle O, Parts L, Piipari M, Winn J, Durbin R (2012) Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 7(3):500–507
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100(16):9440–9445
de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015) MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11(4):e1004219
de Leeuw CA, Neale BM, Heskes T, Posthuma D (2016) The statistical properties of gene-set analysis. Nat Rev Genet 17(6):353–364
de Leeuw CA, Stringer S, Dekkers IA, Heskes T, Posthuma D (2018) Conditional and interaction gene-set analysis reveals novel functional pathways for blood pressure. Nat Commun 9(1):3768
Subramanian A, Tamayo P Fau - Mootha VK, Mootha Vk Fau - Mukherjee S, Mukherjee S, Fau-Ebert BL, Ebert Bl Fau-Gillette MA, Gillette Ma Fau - Paulovich A, et al Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. (0027–8424 (Print)).
Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC et al (2021) Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res 49(D1):D1138-d1143
Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL et al (2015) The Comparative Toxicogenomics Database’s 10th year anniversary: update 2015. Nucleic Acids Res 43(Database issue):D914-920
Nöthen MM, Nieratschker V, Cichon S, Rietschel M (2010) New findings in the genetics of major psychoses. Dialogues Clin Neurosci 12(1):85–93
Dai Y, O’Brien TD, Pei G, Zhao Z, Jia P (2020) Characterization of genome-wide association study data reveals spatiotemporal heterogeneity of mental disorders. BMC Med Genom 13(Suppl 11):192
Brucato N, Guadalupe T, Franke B, Fisher SE, Francks C (2015) A schizophrenia-associated HLA locus affects thalamus volume and asymmetry. Brain Behav Immun 46:311–318
Seshasubramanian V, Raghavan V, SathishKannan AD, Naganathan C, Ramachandran A, Arasu P et al (2020) Association of HLA-A, -B, -C, -DRB1 and -DQB1 alleles at amino acid level in individuals with schizophrenia: a study from South India. Int J Immunogenet 47(6):501–511
Bahi-Buisson N, Poirier K, Fourniol F, Saillour Y, Valence S, Lebrun N et al (2014) The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain 137(Pt 6):1676–1700
Hammerschlag AR, Byrne EM, Agbessi M, Ahsan H, Alves I, Andiappan A et al (2020) Refining attention-deficit/hyperactivity disorder and autism spectrum disorder genetic loci by integrating summary data from genome-wide association, gene expression, and DNA methylation studies. Biol Psychiat 88(6):470–479
Pain O, Pocklington AJ, Holmans PA, Bray NJ, O’Brien HE, Hall LS et al (2019) Novel insight into the etiology of autism spectrum disorder gained by integrating expression data with genome-wide association statistics. Biol Psychiatry 86(4):265–273
van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA et al (2016) Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry 21(4):585
Chen LP, Dai HY, Dai ZZ, Xu CT, Wu RH (2014) Anterior cingulate cortex and cerebellar hemisphere neurometabolite changes in depression treatment: A 1H magnetic resonance spectroscopy study. Psychiatry Clin Neurosci 68(5):357–364
Avino T, Hutsler JJ (2021) Supernumerary neurons within the cerebral cortical subplate in autism spectrum disorders. Brain Res 1760:147350
Mohammadi N, Asle-Rousta M, Rahnema M, Amini R (2021) Morin attenuates memory deficits in a rat model of Alzheimer’s disease by ameliorating oxidative stress and neuroinflammation. Eur J Pharmacol 910:174506
Ben-Azu B, Aderibigbe AO, Omogbiya IA, Ajayi AM, Iwalewa EO Morin pretreatment attenuates schizophrenia-like behaviors in experimental animal models. (2194–9387 (Electronic))
Deng L, Zhou X, Tao G, Hao W, Wang L, Lan Z et al (2022) Ferulic acid and feruloylated oligosaccharides alleviate anxiety and depression symptom via regulating gut microbiome and microbial metabolism. Food Res Int 162(Pt A):111887
Shoeb A, Chowta M, Pallempati G, Rai A, Singh A (2013) Evaluation of antidepressant activity of vanillin in mice. Indian J Pharmacol 45(2):141–144
Xu J, Xu H, Liu Y, He H, Li G (2015) Vanillin-induced amelioration of depression-like behaviors in rats by modulating monoamine neurotransmitters in the brain. Psychiatry Res 225(3):509–514
Wang L, Sun J, Miao Z, Jiang X, Zheng Y, Yang G (2022) Quercitrin improved cognitive impairment through inhibiting inflammation induced by microglia in Alzheimer’s disease mice. NeuroReport 33(8):327–335
Peng Y, Xu D, Ding Y, Zhou X (2022) Supplementation of PQQ from pregnancy prevents MK-801-induced schizophrenia-like behaviors in mice. Psychopharmacology 239(7):2263–2275
Navabi SP, Sarkaki A, Mansouri E, Badavi M, Ghadiri A, Farbood Y (2018) The effects of betulinic acid on neurobehavioral activity, electrophysiology and histological changes in an animal model of the Alzheimer’s disease. Behav Brain Res 337:99–106
Machado DG, Cunha MP, Neis VB, Balen GO, Colla A, Bettio LEB et al (2013) Antidepressant-like effects of fractions, essential oil, carnosol and betulinic acid isolated from Rosmarinus officinalis L. Food Chem 136(2):999–1005
Acknowledgements
This study was conducted using the UK Biobank Resource (Application 46478).
Funding
This work was supported by The National Natural Science Foundation of China (82273753) and The Natural Science Basic Research Plan in Shaanxi Province of China (2021JCW-08).
Author information
Authors and Affiliations
Contributions
SS had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. HZ contributed equally to the work as co-senior authors. YW and FZ conceptualized and designed the study. All authors contributed in acquisition, analysis, and interpretation of the data. SS drafted the manuscript. YW, HZ helped with critical revision of the manuscript for important intellectual content. YW and XC performed statistical analysis. Yan Wen and Yumeng Jia provided administrative, technical, or material support. Yan Wen supervised the study.
Corresponding author
Ethics declarations
Conflicts of interest
All authors report no biomedical financial interests or potential conflicts of interest.
Institutional review board statement
This study has been approved by UKB (Application 46478) and obtained participants' health-related records.
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, S., Zhang, H., Chu, X. et al. Identifying novel chemical-related susceptibility genes for five psychiatric disorders through integrating genome-wide association study and tissue-specific 3′aQTL annotation datasets. Eur Arch Psychiatry Clin Neurosci (2024). https://doi.org/10.1007/s00406-023-01753-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00406-023-01753-0