Abstract
Background
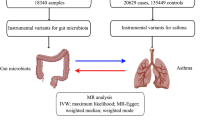
The nasal cavity and gut are interconnected, both housing a rich natural microbiome. Gut microbiota may interact with nasal microbiota and contribute to the development of chronic rhinosinusitis (CRS). However, the specific role of gut microbiota in CRS has not been fully investigated. Therefore, we conducted a two-sample Mendelian randomization study to reveal the potential genetic causal effect of gut microbiota on CRS.
Methods
We performed a two-sample Mendelian Randomization (MR) analysis using aggregated data from genome-wide association studies (GWAS) on gut microbiota and CRS. The primary method used to assess the causal relationship between gut microbiota and CRS was the inverse variance weighting (IVW) method. In addition, sensitivity analyses were conducted to evaluate the robustness of the MR results, including heterogeneity, pleiotropy, and leave-one-out tests.
Results
Genetically predicted twelve gut microbiota, including class Coriobacteriia, class Methanobacteria, family Coriobacteriaceae, family Methanobacteriaceae, family Pasteurellaceae, genus Haemophilus, genus Ruminococcus torques group, genus Subdoligranulum, order Coriobacteriales, order Methanobacteriales, order Pasteurellales, and phylum Proteobacteria, demonstrated a potential inhibitory effect on CRS risk (P < 0.05). In addition, four gut microbiota, including family Streptococcaceae, genus Clostridium innocuum group, genus Oscillospira, and genus Ruminococcaceae NK4A214 group, exhibited a causal role in increasing CRS risk (P < 0.05). Sensitivity analyses showed no evidence of heterogeneity or pleiotropy (P > 0.05).
Conclusions
This study reveals the causal relationship between specific gut microbiota and CRS, which provides a new direction and theoretical foundation for the future development of interventions and prevention and treatment strategies for CRS.


Similar content being viewed by others
Availability of data and materials
Publicly available data sets were analyzed in this study. The data of gut microbiota and CRS (ID: ebi-a-GCST90018823) can be obtained from https://gwas.mrcieu.ac.uk/.
References
Fokkens WJ et al (2020) European position paper on rhinosinusitis and nasal polyps 2020. Rhinology 58(Suppl S29):1–464
Van Crombruggen K et al (2011) Pathogenesis of chronic rhinosinusitis: inflammation. J Allergy Clin Immunol 128(4):728–732
McLean MH et al (2015) Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut 64(2):332–341
Al Bander Z et al (2020) The gut microbiota and inflammation: an overview. Int J Environ Res Public Health 17(20):7618
Huang J et al (2020) Analysis of intestinal flora in patients with chronic rhinosinusitis based on highthroughput sequencing. Nan Fang Yi Ke Da Xue Xue Bao 40(9):1319–1324
Davies NM, Holmes MV, Davey Smith G (2018) Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362:601
Swanson SA et al (2017) Nature as a trialist? Deconstructing the analogy between Mendelian randomization and randomized trials. Epidemiology 28(5):653–659
Sekula P et al (2016) Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol 27(11):3253–3265
Kurilshikov A et al (2021) Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet 53(2):156–165
Sakaue S et al (2021) A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet 53(10):1415–1424
Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37(7):658–665
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44(2):512–525
Bowden J et al (2016) Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40(4):304–314
Hemani G et al (2018) The MR-Base platform supports systematic causal inference across the human phenome. Elife 7:e34408
Nishio J, Honda K (2012) Immunoregulation by the gut microbiota. Cell Mol Life Sci 69(21):3635–3650
Cox AJ, West NP, Cripps AW (2015) Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol 3(3):207–215
Kostic AD, Xavier RJ, Gevers D (2014) The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146(6):1489–1499
Huang R et al (2022) Effects of intestinal microbes on rheumatic diseases: a bibliometric analysis. Front Microbiol 13:1074003
Gurung M et al (2020) Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 51:102590
de Vos WM et al (2022) Gut microbiome and health: mechanistic insights. Gut 71(5):1020–1032
Bassis CM et al (2014) The nasal cavity microbiota of healthy adults. Microbiome 2:27
Abbas EE et al (2021) Distinct clinical pathology and microbiota in chronic rhinosinusitis with nasal polyps endotypes. Laryngoscope 131(1):E34-e44
Binda C et al (2018) Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig Liver Dis 50(5):421–428
Gao B, Gupta RS (2012) Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev 76(1):66–112
Sarveswari HB, Solomon AP (2019) Profile of the Intervention potential of the phylum Actinobacteria toward quorum sensing and other microbial virulence strategies. Front Microbiol 10:2073
Shi J et al (2021) Sex-specific associations between gut microbiome and non-alcoholic fatty liver disease among urban Chinese adults. Microorganisms 9(10):2118
Macfarlane S, Macfarlane GT (2003) Regulation of short-chain fatty acid production. Proc Nutr Soc 62(1):67–72
Schirmer M et al (2016) Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167(4):1125-1136.e8
Dorrestein PC, Mazmanian SK, Knight R (2014) Finding the missing links among metabolites, microbes, and the host. Immunity 40(6):824–832
Cavaglieri CR et al (2003) Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci 73(13):1683–1690
Klampfer L et al (2003) Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol Cancer Res 1(11):855–862
Smith LD, King E (1962) Clostridium innocuum, sp. n., a sporeforming anaerobe isolated from human infections. J Bacteriol 83(4):938–939
Cherny KE et al (2021) Clostridium innocuum: microbiological and clinical characteristics of a potential emerging pathogen. Anaerobe 71:102418
Acknowledgements
We thank all the genetics consortiums for making the GWAS summary data publicly available.
Funding
This work was supported by the Natural Science Foundation of Sichuan Province (Grant Number 2023NSFSC0621) and Chengdu Medical Research Project (Grant Number 2022013).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent
This study only used publicly available GWAS summary data, so ethical approval is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, F., Cai, B., Luo, J. et al. Gut microbiota and chronic rhinosinusitis: a two-sample Mendelian randomization study. Eur Arch Otorhinolaryngol 281, 3025–3030 (2024). https://doi.org/10.1007/s00405-024-08468-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-024-08468-5