Abstract
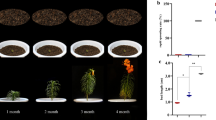
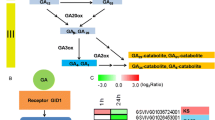
The long-followed practice of using garlic cloves for garlic asexual reproduction has resulted in virus accumulation and genetic depression. Aerial bulbs, the preferred material for virus-free garlic seedling cultivation in vitro, undergo dormancy, but gibberellin can quickly break this state. In this study, exogenous GA3 was applied to dormant aerial bulbs and transcriptome sequencing was then performed using control and gibberellin-treated aerial bulbs. Genes differentially expressed in response to the gibberellin treatment were analyzed and screened and their relationship to GA3-induced dormancy break of aerial bulbs was explored. According to our results, exogenous GA3 was able to effectively break aerial bulb dormancy. Transcriptome sequencing uncovered 4931 and 74,636 genes that were respectively up- and down-regulated in response to gibberellin treatment. 37,957 differentially expressed genes were successfully assigned to KEGG pathways, among which the most DEGs were annotated in metabolism pathway (23,245 genes, accounting for 61.24%), followed by the genetic information processing pathway (9235 genes, accounting for 24.33%). More DEGs were enriched in spliceosome, ribosome, ribosome biogenesis in eukaryotes, and ubiquitin-mediated proteolysis pathways and the enrichment results were significant. In addition, carbohydrate and amino acid metabolism pathways are active. The significant DEGs related to calcium signaling, hormonal signaling, and transcription factors were screened out. Among them, the expression of DEGs related to hormonal signaling was consistent with the change of endogenous hormone content in aerial bulbs. Finally, the outcome of a qRT-PCR analysis was consistent with our transcriptome sequencing results, thus indicating that the latter was reliable.







Similar content being viewed by others
References
Altschul SF (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bai X, Zhai H, Zhu YM (2011) Arabidopsis transcription factor MYBC1 mediates seed germination by ABA. J Northeast Agr U 42:118–124
Belin C, Megies C, Hauserová E, Lopezmolina L (2009) Abscisic acid represses growth of the arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21:2253–2268
Boulikas T (2010) Putative nuclear localization signals (NLS) in protein transcription factors. J Cell Biochem 55:32–58
Conesa A, Terol J, Robles M (2005) Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Dong YH, Gu QY, Yuan L, Wang LX, Sun XD, Liu SQ (2019) Rapid propagation of virus-free garlic aerial bulbs. J Plant Physiol 55:1315–1324
Dong YH, Wang LX, Gu QY, Guan MJ, Dong F, Sun XD, Liu SQ (2020) Effects of low temperature and exogenous GA3 on releasing aerial bulb dormancy in garlic and its physiological and biochemical mechanisms. J Plant Physiol 56:256–264
Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX (2009) Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry-Moscow 74:1–11
Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18:53–63
Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11:1897–1909
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Xian A, Lin F, Raychowdhury R, Zeng Q (2011) Trinity: reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat Biotechnol 29:644–652
Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160:83–92
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20:735–747
Hepler PK (2005) Calcium: a central regulator of plant growth and development. Plant Cell 17:2142–2155
Hou HL, Zhang CW, Hou XL (2020) Cloning and functional analysis of BcMYB101 gene involved in leaf development in Pak Choi (Brassica rapa ssp, Chinensis). Int J Mol Sci 21:2750
Huang XZ, Jiang CF, Liao LL (2006) Progress on molecular foundation of GA biosynthesis pathway and signaling. Chin Bull Bot 23:499–510
Huang T, Yang JL, Yu D, Han XY, Wang XQ (2020) Bioinformatics analysis of WRKY transcription factors in grape and their potential roles prediction in sugar and abscisic acid signaling pathway. J Plant Biochem Biot. https://doi.org/10.1007/s13562-020-00571-y
Jian LC, Deng JM, Li JH, Li PH (2003) Seasonal alteration of the cytosolic and nuclear Ca2+ concentrations in overwintering woody and herbaceous perennials in relation to the development of dormancy and cold hardiness. J Am Soc Hortic Sci 128:29–35
Kamenetsky R, Shafir IL, Baizerman M, Khassanov F, Kik C, Rabinowitch HD (2004) Garlic (Allium sativum L.) and its wild relatives from Central Asia: evaluation for fertility potential. Acta Horticul 637:83–91
Kucera B, Cohn MA, Leubnermetzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15:281–307
Leubner-Metzger G (2003) Functions and regulation of beta-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci Res 13:17–34
Li WG, Xu RR, Yan XG, Liang DM, Zhang L, Qin XY, Caiyin Q, Zhao GR, Xiao WH, Hu ZN, Qiao JJ (2019) De novo leaf and root transcriptome analysis to explore biosynthetic pathway of Celangulin V in Celastrus angulatus maxim. BMC Genomics 20:7
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2010) Seed dormancy and ABA metabolism in arabidopsis and barley: the role of ABA 8’-hydroxylase. Plant J 45:942–954
Nambara E, Marion-Poll A (2003) ABA action and interactions in seeds. Trends Plant Sci 8:217
Pang XQ, Halaly T, Crane O, Keilin T, Keren A, Ogrodovitch A, Galbraith D, Or E (2007) Involvement of calcium signalling in dormancy release of grape buds. J Exp Bot 58:3249–3262
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B (2003) TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652
Sathyanarayanan PV, Poovaiah BW (2004) Decoding Ca2+ signals in plants. Crit Rev Plant Sci 23:1–11
Shi CY (2011) Deep sequencing of the camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12:131
Sun XD, Zhou SM, Meng FL, Liu SQ (2012) De novo assembly and characterization of the garlic (Allium Sativum) bud transcriptome by Illumina sequencing. Plant Cell Rep 31:1823–1828
Sun XD, Ren YQ, Zhang XZ, Lian HF, Zhou SM, Liu SQ (2016) Overexpression of a garlic nuclear factor Y (NF-Y) B gene, AsNF-YB3, affects seed germination and plant growth in transgenic tobacco. Plant Cell Tiss Org 127:513–523
Tan Y, Gao DS, Li L, Wei HR, Wang JW, Liu QZ (2015) Relationships between H2O2 metabolism and Ca2+ transport in dormancy-breaking process of nectarine floral buds. Chin J Appl Ecol 26:425–429
Torres TT, Metta M, Ottenwälder B, Schlötterer C (2010) Gene expression profiling by massively parallel sequencing. Genome Res 18:172
Tuan PA, Kumar R, Rehal PK, Toora P, Ayele BT (2018) Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front Plant Sci 9:668
Wan JP, Wang RT, Wang RL, Ju Q, Wang YB, Xu J (2019) Comparative physiological and transcriptomic analyses reveal the toxic effects of ZnO nanoparticles on plant growth. Environ Sci Technol 53:4235–4244
Wang CM, Wang HW, Zhang JS, Chen SY (2008) A seed-specific AP2-domain transcription factor from soybean plays a certain role in regulation of seed germination. Sci China Life Sci 4:336–345
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an r package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Wang Y, Chu YJ, Xue HW (2012) Inositol polyphosphate 5-phosphatase-controlled Ins(1,4,5)P3/Ca2+ is crucial for maintaining pollen dormancy and regulating early germination of pollen. Development 139:2221–2233
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:293–297
Yue C, Cao HL, Hao XY, Zeng JM, Qian WJ, Guo YQ, Ye NX, Yang YJ, Wang XC (2018) Differential expression of gibberellin- and abscisic acid-related genes implies their roles in the bud activity-dormancy transition of tea plants. Plant Cell Rep 37:425–441
Zhang SZ, Li JR (2006) Analysis of factors affecting the germination of bulbils in the inflorescence of garlic (Allium sativum L.). J Sichuan Agr Univ 24:148–151
Zheng CL, Acheampong AK, Shi ZW, Kamiya YJ (2018) Distinct gibberellin functions during and after grapevine bud dormancy release. J Exp Bot. https://doi.org/10.1093/jxb/ery022
Acknowledgements
This research was funded by the Shandong Provincial Natural Science Foundation (Grant No. ZR2016CM33) and the Opening Foundation of Key laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture (Grant No. IVF201805).
Author information
Authors and Affiliations
Contributions
YD and HF were responsible for collecting tissue samples. YH and YZ performed the RNA extractions and preparations. YD, HF, and YZ analyzed the data. YD, XS, and SL contributed to the writing and revising of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: Theo Lange.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, Y., Fang, H., Hou, Y. et al. Transcriptome Analysis Reveals Differential Gene Expression in Garlic Aerial Bulbs in Response to Gibberellin Application. J Plant Growth Regul 41, 2967–2979 (2022). https://doi.org/10.1007/s00344-021-10488-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10488-y